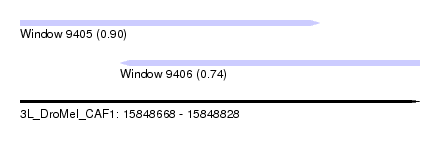
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,848,668 – 15,848,828 |
| Length | 160 |
| Max. P | 0.898258 |

| Location | 15,848,668 – 15,848,788 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.91 |
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -21.06 |
| Energy contribution | -21.54 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15848668 120 + 23771897 AACUGCUAAGCAAUUUAAUCGGAUAAAAACUAUUUCGAAAUGUAUGCAACAAAGUGAGCAUCGCAUUUCGCGGUAGAGUGUGUGCUUUUAUGUGGCUUCUAUUCGCAUAAAUUUAUUAAU ....((...(((.((((......))))..((((..((((((((((((..........)))).))))))))..))))..)))..)).(((((((((.......)))))))))......... ( -26.20) >DroSec_CAF1 55889 120 + 1 AACUGCUAAGCAAUUUAAUCGGAUAAAAACUAUUCCGAAAUUUAUGCAACAAAGUGCGCAUCGCAUUUCGCGGUAGAGAGUGUGCUUUUAUGUGGCUUCUAGCCGCAUAAAUUUAUUAAU .(((((...(((......(((((..........)))))......)))....(((((((...))))))).))))).......(((..((((((((((.....))))))))))..))).... ( -33.70) >DroSim_CAF1 57115 120 + 1 AACUGCUAAGCAAUUUAAUCGGAUAAAAACUAUUUCGAAAUUUAUGCAACAAAGUGUGCAUCGCAUUUCGCGGUAGAGUGUGUGCUUUUAUGUGGCUUCUAGCCGCAUAAAUUUAUUAAU ....((...(((.((((......))))..((((..((((((..(((((........)))))...))))))..))))..)))..)).((((((((((.....))))))))))......... ( -29.50) >DroEre_CAF1 55064 115 + 1 AACUACUAAGCAUUAUAAUCGGAUAA-AACUAUUCCAAAAUGUAUGCAACAAA----GUAUCGCAUUUCGCGGUAGAGCACUCGCUUUUAUGCGGCUUCUAUCCACAUCAAUUUAUUAAU ....................(((((.-..((((..(.((((((((((......----)))).)))))).)..))))(((...(((......))))))..)))))................ ( -21.20) >DroYak_CAF1 97201 119 + 1 AACUACUAAGCAUUAUAAUCGGAUAA-AACUAUUCCAAAAUGUAUGCAACAAAAUGAGCAUCGCAUUUCGCGGUAGAGCACACGCUUUUAUGUGGCUUUUAUCCACAUCAAUUUAUUAAU ....................((((((-(.((((..(.((((((((((..........)))).)))))).)..))))(((.((((......))))))))))))))................ ( -24.70) >consensus AACUGCUAAGCAAUUUAAUCGGAUAAAAACUAUUCCGAAAUGUAUGCAACAAAGUGAGCAUCGCAUUUCGCGGUAGAGCGUGUGCUUUUAUGUGGCUUCUAUCCGCAUAAAUUUAUUAAU ..(((((..((.........(((..........)))(((((((((((..........)))).))))))))))))))..........(((((((((.......)))))))))......... (-21.06 = -21.54 + 0.48)


| Location | 15,848,708 – 15,848,828 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.73 |
| Mean single sequence MFE | -28.36 |
| Consensus MFE | -22.20 |
| Energy contribution | -23.24 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15848708 120 - 23771897 CCCGCGGUGAAUAUUGACUGAAAUACCGGCCAAAACGGCCAUUAAUAAAUUUAUGCGAAUAGAAGCCACAUAAAAGCACACACUCUACCGCGAAAUGCGAUGCUCACUUUGUUGCAUACA ..(((((((..................((((.....)))).........((((((.(.........).))))))...........)))))))..(((((((.........)))))))... ( -27.70) >DroSec_CAF1 55929 120 - 1 CCCGCGGUGAAUAUUGACUGAAAUACCGGCAAAAACGGCCAUUAAUAAAUUUAUGCGGCUAGAAGCCACAUAAAAGCACACUCUCUACCGCGAAAUGCGAUGCGCACUUUGUUGCAUAAA ..(((((((..................(((.......))).........((((((.(((.....))).))))))...........)))))))..(((((((.........)))))))... ( -31.70) >DroSim_CAF1 57155 120 - 1 CCCGCGGUGAAUAUUGACUGAAAUACCGGCAAAAACGGCCAUUAAUAAAUUUAUGCGGCUAGAAGCCACAUAAAAGCACACACUCUACCGCGAAAUGCGAUGCACACUUUGUUGCAUAAA ..(((((((..................(((.......))).........((((((.(((.....))).))))))...........)))))))..(((((((.........)))))))... ( -31.70) >DroEre_CAF1 55103 114 - 1 CCCGAGGUAAA--UUGACUGAAAUACCGACAAAAACUGCCAUUAAUAAAUUGAUGUGGAUAGAAGCCGCAUAAAAGCGAGUGCUCUACCGCGAAAUGCGAUAC----UUUGUUGCAUACA .....((((..--..........))))..........................(((((.((((.(((((......))).))..)))))))))..(((((((..----...)))))))... ( -23.60) >DroYak_CAF1 97240 118 - 1 CCCGAGGUAAA--UUGACUGAAAUACCGGCAAAAACUGCCAUUAAUAAAUUGAUGUGGAUAAAAGCCACAUAAAAGCGUGUGCUCUACCGCGAAAUGCGAUGCUCAUUUUGUUGCAUACA ..(((((((..--..........))))((((.....)))).........)))((((((.......))))))......((((((......((((((((.......)))))))).)))))). ( -27.10) >consensus CCCGCGGUGAAUAUUGACUGAAAUACCGGCAAAAACGGCCAUUAAUAAAUUUAUGCGGAUAGAAGCCACAUAAAAGCACACACUCUACCGCGAAAUGCGAUGCUCACUUUGUUGCAUACA ..(((((((..................((((.....)))).........((((((.((.......)).))))))...........)))))))..((((((((.......))))))))... (-22.20 = -23.24 + 1.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:20 2006