
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,844,342 – 15,844,487 |
| Length | 145 |
| Max. P | 0.880613 |

| Location | 15,844,342 – 15,844,447 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.09 |
| Mean single sequence MFE | -17.10 |
| Consensus MFE | -11.82 |
| Energy contribution | -11.38 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15844342 105 + 23771897 -AAUAUU--------------UGUUUUACUCAUUUUUACCUUAUUGAUUUAAGAACAUAUUCAGUAUUUUCUCUUCCUGUAAGGUACUUCGUGUUUGUGCUUUGCAUUUUUUAUAGCCAG -......--------------....................((((((.............))))))...........((((((((((.........)))))))))).............. ( -13.12) >DroSec_CAF1 51436 105 + 1 -AAUAUU--------------UCUUUUACCCAUUUUAACCUAAAUGAUUUUAACAAAUAUUCAGUAUUUUCUCUUCCUGUAAGGCACUUCGUAUUUGUGCUUUGCAUUUUUUAUGGCCAG -((((((--------------(.((.....(((((......))))).....)).)))))))................((((((((((.........)))))))))).............. ( -15.60) >DroSim_CAF1 52657 105 + 1 -AAUAUU--------------UCUUUUACCCAUUUUAACCUUAAUGAUUUUGAGAAAUAUUCUGUAUUUUCUCUUCCUGUAAGGCACUUCGUAUUUGUGCUUUGCAUUUUUUAUGGCCAG -((((((--------------((((.....((((........)))).....))))))))))................((((((((((.........)))))))))).............. ( -21.10) >DroEre_CAF1 50560 115 + 1 AAAUAUUCAGCUUUCCAAAUUCGUUGAACUCAGUCUAACCGCAA----UUUAAUAAUUGUGCACACUUUUCUCUUCGUGUAGAGCACUGCUUGUUCGUGCUUUGCAUUUU-UAUGUUCCG .....((((((...........))))))...........(((((----((....)))))))...............(((((((((((.........)))))))))))...-......... ( -22.00) >DroYak_CAF1 92834 110 + 1 AAAUGUUCAGGUUUCUAAAUU---------UAGUUUAAUUGCAAUAAUUUUAAUAACUAUUCAGACUUUU-UCUUUCUGUAGAGCUCUGCUUGUUCGUGCUUUGCAUUUUGUAUGCCCUG .......((((..........---------..........(((..................((((.....-....))))..((((.......)))).)))...((((.....)))))))) ( -13.70) >consensus _AAUAUU______________UCUUUUACCCAUUUUAACCUCAAUGAUUUUAAUAAAUAUUCAGUAUUUUCUCUUCCUGUAAGGCACUUCGUGUUUGUGCUUUGCAUUUUUUAUGGCCAG .............................................................................((((((((((.........)))))))))).............. (-11.82 = -11.38 + -0.44)

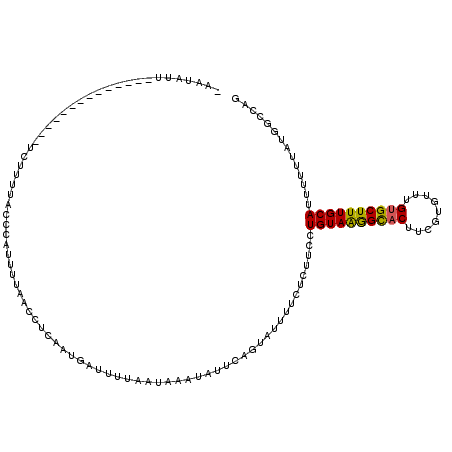

| Location | 15,844,367 – 15,844,487 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.50 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -16.72 |
| Energy contribution | -18.12 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15844367 120 + 23771897 UUAUUGAUUUAAGAACAUAUUCAGUAUUUUCUCUUCCUGUAAGGUACUUCGUGUUUGUGCUUUGCAUUUUUUAUAGCCAGUACUUUAUUUUUAUGUAUUUUCCGUAUGAGAUUCUUUGAA ...................(((((.....((((....((((((((((.........)))))))))).....((..(..(((((...........)))))..)..)).))))....))))) ( -18.20) >DroSec_CAF1 51461 120 + 1 UAAAUGAUUUUAACAAAUAUUCAGUAUUUUCUCUUCCUGUAAGGCACUUCGUAUUUGUGCUUUGCAUUUUUUAUGGCCAGUACUUUAUUUUUAUGUAUUUUCCGUAUGAGAUUCUUUGAA ...................(((((.....((((....((((((((((.........)))))))))).....(((((..(((((...........)))))..))))).))))....))))) ( -25.80) >DroSim_CAF1 52682 120 + 1 UUAAUGAUUUUGAGAAAUAUUCUGUAUUUUCUCUUCCUGUAAGGCACUUCGUAUUUGUGCUUUGCAUUUUUUAUGGCCAGUACUUUAUUUUUAUGUAUUUUCCGUAUGAGAUUUUUUGAA ((((.((((((((((((..........))))))....((((((((((.........)))))))))).....(((((..(((((...........)))))..))))).))))))..)))). ( -25.70) >DroEre_CAF1 50600 115 + 1 GCAA----UUUAAUAAUUGUGCACACUUUUCUCUUCGUGUAGAGCACUGCUUGUUCGUGCUUUGCAUUUU-UAUGUUCCGUACUUUAUUUUUUAGUAUUUCCCGUGUGAGAUUCUUUGAA ((((----((....)))))).(((((..........(((((((((((.........)))))))))))...-........(((((.........))))).....)))))............ ( -24.60) >DroYak_CAF1 92865 119 + 1 GCAAUAAUUUUAAUAACUAUUCAGACUUUU-UCUUUCUGUAGAGCUCUGCUUGUUCGUGCUUUGCAUUUUGUAUGCCCUGUACUUUAUUUUUAAGAAUUUCCCGUAUGAGAUUCUUUGAA .....................((((.....-....))))..((((.......))))((((...((((.....))))...)))).....(((.(((((((((......))))))))).))) ( -19.60) >consensus UCAAUGAUUUUAAUAAAUAUUCAGUAUUUUCUCUUCCUGUAAGGCACUUCGUGUUUGUGCUUUGCAUUUUUUAUGGCCAGUACUUUAUUUUUAUGUAUUUUCCGUAUGAGAUUCUUUGAA ...................(((((.....((((....((((((((((.........)))))))))).....(((((..(((((...........)))))..))))).))))....))))) (-16.72 = -18.12 + 1.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:16 2006