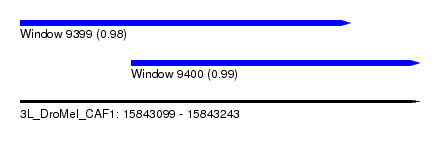
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,843,099 – 15,843,243 |
| Length | 144 |
| Max. P | 0.987596 |

| Location | 15,843,099 – 15,843,218 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 92.27 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -28.06 |
| Energy contribution | -28.46 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
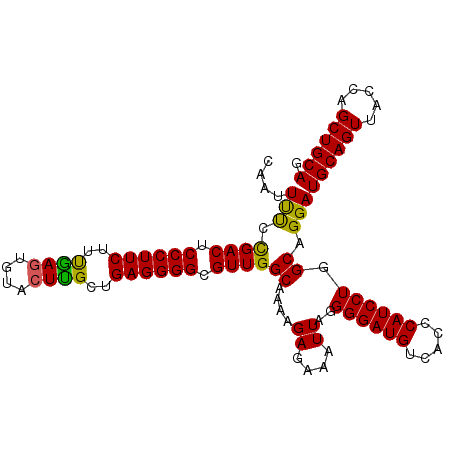
>3L_DroMel_CAF1 15843099 119 + 23771897 UGUUCUUAAUCUUAAAAAACAACUUAAUCUUUCGAAAUGGCAAUUUUCCGACUCCCUUCUUUGAGUGUACUCGCUGAGGGGCGUUGGCAAAAGAGAAAUUAGGGGAUGUCACCCAUCCU ......................((((((.(((((((((....)))))(((((.((((((..((((....))))..)))))).))))).....)))).))))))(((((.....))))). ( -34.80) >DroSec_CAF1 50186 119 + 1 UGUUCUUAAUCUUAAAAAACAACUUAAUCUUUCGAAAUGGCAAUUCUCCGACUCCCUUCUUCAAGUGUACUUGCUGAGGGGCGUUGGCCAAAGAGAAAUUAGGGGAUGUCACCCAUCCU ......................((((((.((((....((((.......((((.((((((..((((....))))..)))))).))))))))..)))).))))))(((((.....))))). ( -35.21) >DroSim_CAF1 51401 119 + 1 UGCUCUUAAUCUUAAAAAACAACUUAAUCUUUCGAAAUGGCAAUUCUCCGACUCCCUUCUUCGAGUGUACUUGCUGAGGGGCGUUGGCAAAAGAGAAAUUAGGGGAUGUCACCCAUCCU ......................((((((.(((((((.......))).(((((.((((((..((((....))))..)))))).))))).....)))).))))))(((((.....))))). ( -33.60) >DroEre_CAF1 49326 119 + 1 UGUUCUUGAUCUUAAAAAACAACUUAAUCUUUCGAAAUAGCCAUUUCGUGACUCCCUUCUCUGAGUGUACUUGCUGAGGGGCGUUGGCAAAAGAGAAAUUAGGGGAUGUCACCCAUCCU ((((.((.......)).)))).((((((.(((((((((....)))))..(((.((((((..((((....))))..)))))).))).......)))).))))))(((((.....))))). ( -29.50) >DroYak_CAF1 91560 119 + 1 UGUUUUGGAUCUUAAAAAACAACUUAAUCUUUAAAAAUAGCAAUUUCGUGACUCCCUUCGUUAAGUGUAGUUGCUGAGGGGCGUUGGCAAAAGAGAAAUUAGGGGAUGUCACCCAUCCU ((((((.........)))))).((((((((((.......((......))(((.(((((((.(((......))).))))))).)))......)))))..)))))(((((.....))))). ( -24.10) >consensus UGUUCUUAAUCUUAAAAAACAACUUAAUCUUUCGAAAUGGCAAUUUUCCGACUCCCUUCUUUGAGUGUACUUGCUGAGGGGCGUUGGCAAAAGAGAAAUUAGGGGAUGUCACCCAUCCU ......................((((((.(((((((((....)))))(((((.((((((..((((....))))..)))))).))))).....)))).))))))(((((.....))))). (-28.06 = -28.46 + 0.40)


| Location | 15,843,139 – 15,843,243 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 94.23 |
| Mean single sequence MFE | -36.71 |
| Consensus MFE | -32.98 |
| Energy contribution | -33.10 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15843139 104 + 23771897 CAAUUUUCCGACUCCCUUCUUUGAGUGUACUCGCUGAGGGGCGUUGGCAAAAGAGAAAUUAGGGGAUGUCACCCAUCCUGGCAGGAUGCAGUUACCAGCUGCAG ...(((((((((.((((((..((((....))))..)))))).))))).)))).........(((.......)))((((.....))))(((((.....))))).. ( -38.30) >DroSec_CAF1 50226 104 + 1 CAAUUCUCCGACUCCCUUCUUCAAGUGUACUUGCUGAGGGGCGUUGGCCAAAGAGAAAUUAGGGGAUGUCACCCAUCCUGGCAGGAUGCAGUUACCAGCUGCAG ..(((((.((((.((((((..((((....))))..)))))).))))(((...((....))..((((((.....))))))))))))))(((((.....))))).. ( -39.70) >DroSim_CAF1 51441 104 + 1 CAAUUCUCCGACUCCCUUCUUCGAGUGUACUUGCUGAGGGGCGUUGGCAAAAGAGAAAUUAGGGGAUGUCACCCAUCCUGGCAGGAUGCAGUUACCAGCUGCAG ...(((((((((.((((((..((((....))))..)))))).)))))....))))......(((.......)))((((.....))))(((((.....))))).. ( -37.50) >DroEre_CAF1 49366 104 + 1 CCAUUUCGUGACUCCCUUCUCUGAGUGUACUUGCUGAGGGGCGUUGGCAAAAGAGAAAUUAGGGGAUGUCACCCAUCCUGGCAGGAUGCAGUUACCAGCUGCAG .......(((((((((((((((...(((...((((....))))...)))..))))).....))))).)))))..((((.....))))(((((.....))))).. ( -36.70) >DroYak_CAF1 91600 104 + 1 CAAUUUCGUGACUCCCUUCGUUAAGUGUAGUUGCUGAGGGGCGUUGGCAAAAGAGAAAUUAGGGGAUGUCACCCAUCCUGGCAGGAUGCAGUUACCAGCUGCAG .......(((((((((((.(((........((((..(......)..))))......))).)))))).)))))..((((.....))))(((((.....))))).. ( -31.34) >consensus CAAUUUUCCGACUCCCUUCUUUGAGUGUACUUGCUGAGGGGCGUUGGCAAAAGAGAAAUUAGGGGAUGUCACCCAUCCUGGCAGGAUGCAGUUACCAGCUGCAG ...(((.(((((.((((((..((((....))))..)))))).))))).)))..........(((.......)))((((.....))))(((((.....))))).. (-32.98 = -33.10 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:14 2006