| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,841,866 – 15,842,016 |
| Length | 150 |
| Max. P | 0.769699 |

| Location | 15,841,866 – 15,841,956 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -22.81 |
| Consensus MFE | -17.57 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15841866 90 - 23771897 CAACACGCGUGACCAGCAGCAUAAGCCCACCCAACUUCCAUU----UUGCA-ACUGCCACGCCCACAUCCCAUUGGGG-CCGUCAGCCCACCGUGA ...((((((((..(((..(((.....................----.))).-.))).))))).............(((-(.....))))...))). ( -21.75) >DroSec_CAF1 48949 92 - 1 CAACACGCGUGACCAGCAGCAUAAGCCCACCCAACUUCCAUU----UUGCAAACUGCCACGCCCACAUCCCAGUGGCCCCCAUCAGCCCACCGUGA ...((((.(((....((.(((.....................----.))).....(((((............)))))........)).))))))). ( -17.95) >DroEre_CAF1 48109 94 - 1 CAACACGCGUGACCAGCAGCAUAAGCCCACCCAGCUUCCACUUCCAUUGCA-GCUGCCACGCCCACAUCCCAGUGGGG-CCUUCAGCCCACCGUGA ...((((((((..((((.(((.((((.......))))..........))).-)))).))))).(((......)))(((-(.....))))...))). ( -28.20) >DroYak_CAF1 90375 90 - 1 CAACACGCGUGACCAGCAGCAUAAGCCCACCCAACUUUCAUU----UUGCA-ACUGCCACGCCCACAUCCCAGUGGGG-CCUUCAGCCCACCGUGA ...((((((((..(((..(((.....................----.))).-.))).))))).(((......)))(((-(.....))))...))). ( -23.35) >consensus CAACACGCGUGACCAGCAGCAUAAGCCCACCCAACUUCCAUU____UUGCA_ACUGCCACGCCCACAUCCCAGUGGGG_CCUUCAGCCCACCGUGA ...((((((((..(((..(((..........................)))...))).)))))..........(((((.........))))).))). (-17.57 = -18.07 + 0.50)

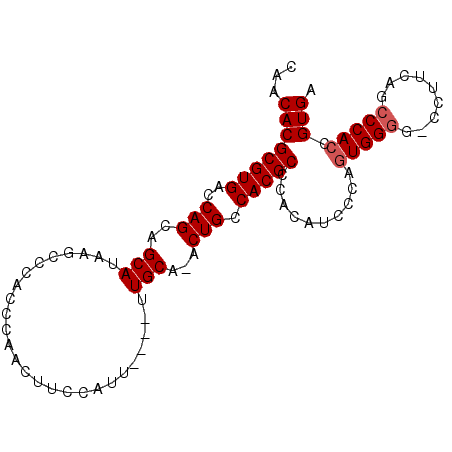

| Location | 15,841,891 – 15,841,993 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -37.39 |
| Energy contribution | -37.33 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.48 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15841891 102 + 23771897 GGAUGUGGGCGUGGCAGU-UGCAA----AAUGGAAGUUGGGUGGGCUUAUGCUGCUGGUCACGCGUGUUGGCCGCGUGUGUCCUGCGCUGAUUUCGCCAUAAUAAAU ...(((((.(((.((...-.))..----.)))(((((..(...(((....)))((.((.((((((((.....)))))))).)).)).)..))))).)))))...... ( -36.50) >DroSec_CAF1 48975 103 + 1 GGAUGUGGGCGUGGCAGUUUGCAA----AAUGGAAGUUGGGUGGGCUUAUGCUGCUGGUCACGCGUGUUGGCCGCGUGUGUCCUGCGCUGAUUUCGCCAUAAUAAAU ((.((..(((......)))..)).----....(((((..(...(((....)))((.((.((((((((.....)))))))).)).)).)..))))).))......... ( -36.60) >DroEre_CAF1 48134 106 + 1 GGAUGUGGGCGUGGCAGC-UGCAAUGGAAGUGGAAGCUGGGUGGGCUUAUGCUGCUGGUCACGCGUGUUGGCCGCGUGUGUCCUGCGCUGAUUUCGCCAUAAUAAAU ((..((.(((((((((((-.((......(((....)))......))....))))))((.((((((((.....)))))))).)).))))).))....))......... ( -39.80) >DroYak_CAF1 90400 102 + 1 GGAUGUGGGCGUGGCAGU-UGCAA----AAUGAAAGUUGGGUGGGCUUAUGCUGCUGGUCACGCGUGUUGGUCGCGUGUGUCCUGCGCUGAUUUCGCCAUAAUAAAU ((..((.(((((((((((-.((..----(((....)))......))....))))))((.((((((((.....)))))))).)).))))).))....))......... ( -36.80) >consensus GGAUGUGGGCGUGGCAGU_UGCAA____AAUGGAAGUUGGGUGGGCUUAUGCUGCUGGUCACGCGUGUUGGCCGCGUGUGUCCUGCGCUGAUUUCGCCAUAAUAAAU ((..((.(((((((((((..((......(((....)))......))....))))))((.((((((((.....)))))))).)).))))).))....))......... (-37.39 = -37.33 + -0.06)

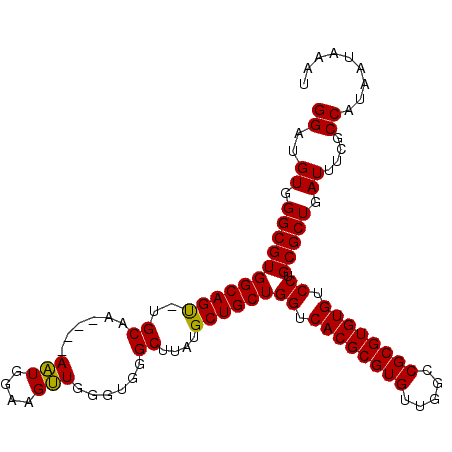

| Location | 15,841,891 – 15,841,993 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -22.77 |
| Energy contribution | -22.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15841891 102 - 23771897 AUUUAUUAUGGCGAAAUCAGCGCAGGACACACGCGGCCAACACGCGUGACCAGCAGCAUAAGCCCACCCAACUUCCAUU----UUGCA-ACUGCCACGCCCACAUCC .........((((....(((.((((((..((((((.......)))))).......((....))..............))----)))).-.)))...))))....... ( -23.60) >DroSec_CAF1 48975 103 - 1 AUUUAUUAUGGCGAAAUCAGCGCAGGACACACGCGGCCAACACGCGUGACCAGCAGCAUAAGCCCACCCAACUUCCAUU----UUGCAAACUGCCACGCCCACAUCC .........((((....(((.((((((..((((((.......)))))).......((....))..............))----))))...)))...))))....... ( -23.20) >DroEre_CAF1 48134 106 - 1 AUUUAUUAUGGCGAAAUCAGCGCAGGACACACGCGGCCAACACGCGUGACCAGCAGCAUAAGCCCACCCAGCUUCCACUUCCAUUGCA-GCUGCCACGCCCACAUCC .........((((....((((((((((..((((((.......))))))...........((((.......)))).....)))..))).-))))...))))....... ( -28.80) >DroYak_CAF1 90400 102 - 1 AUUUAUUAUGGCGAAAUCAGCGCAGGACACACGCGACCAACACGCGUGACCAGCAGCAUAAGCCCACCCAACUUUCAUU----UUGCA-ACUGCCACGCCCACAUCC .........((((....(((.((((((..((((((.......)))))).......((....))..............))----)))).-.)))...))))....... ( -22.30) >consensus AUUUAUUAUGGCGAAAUCAGCGCAGGACACACGCGGCCAACACGCGUGACCAGCAGCAUAAGCCCACCCAACUUCCAUU____UUGCA_ACUGCCACGCCCACAUCC .........((((......((((.((...((((((.......)))))).)).)).))............................((.....))..))))....... (-22.77 = -22.77 + 0.00)

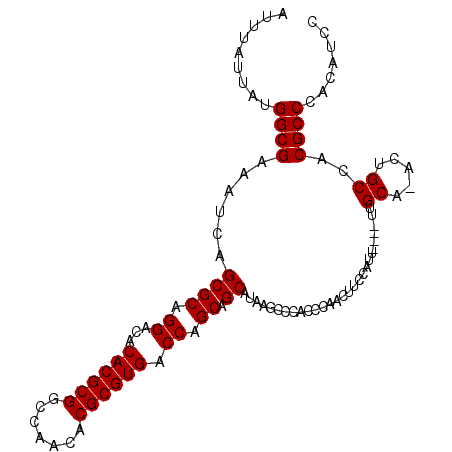

| Location | 15,841,926 – 15,842,016 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 94.49 |
| Mean single sequence MFE | -21.02 |
| Consensus MFE | -18.47 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15841926 90 - 23771897 AGAGAAAUGUCCAAAAAAUGUCAAUUUAUUAUGGCGAAAUCAGCGCAGGACACACGCGGC-----------CAACACGCGUGACCAGCAGCAUAAGCCCAC .......(((((...((((....))))......(((.......))).)))))((((((..-----------.....)))))).......((....)).... ( -20.60) >DroSec_CAF1 49011 90 - 1 AGAGAAAUGUCCAAAAAAUGUCAAUUUAUUAUGGCGAAAUCAGCGCAGGACACACGCGGC-----------CAACACGCGUGACCAGCAGCAUAAGCCCAC .......(((((...((((....))))......(((.......))).)))))((((((..-----------.....)))))).......((....)).... ( -20.60) >DroSim_CAF1 50202 101 - 1 AGAGAAAUGUCCAAAAAAUGUCAAUUUAUUAUGGCGAAAUCAGCGCAGGACACACGCGGCCAACACGCGGCCAACACGCGUGACCAGCAGCAUAAGCCCAC .......(((((...((((....))))......(((.......))).)))))...((.((...((((((.......))))))....)).)).......... ( -24.40) >DroEre_CAF1 48173 90 - 1 AGAGAAACGUCCAAAAAAUGUCAAUUUAUUAUGGCGAAAUCAGCGCAGGACACACGCGGC-----------CAACACGCGUGACCAGCAGCAUAAGCCCAC ........((((...((((....))))......(((.......))).)))).((((((..-----------.....)))))).......((....)).... ( -20.20) >DroYak_CAF1 90435 90 - 1 AGAGAAAUGUCCAAAAAAUGUCAAUUUAUUAUGGCGAAAUCAGCGCAGGACACACGCGAC-----------CAACACGCGUGACCAGCAGCAUAAGCCCAC .......(((((...((((....))))......(((.......))).)))))((((((..-----------.....)))))).......((....)).... ( -19.30) >consensus AGAGAAAUGUCCAAAAAAUGUCAAUUUAUUAUGGCGAAAUCAGCGCAGGACACACGCGGC___________CAACACGCGUGACCAGCAGCAUAAGCCCAC ........((((...((((....))))......(((.......))).)))).((((((..................)))))).......((....)).... (-18.47 = -18.47 + 0.00)

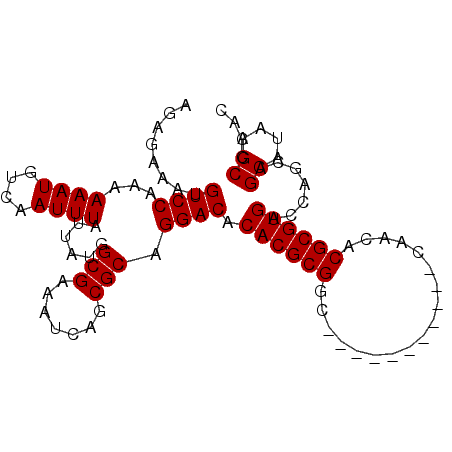
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:11 2006