| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,836,015 – 15,836,119 |
| Length | 104 |
| Max. P | 0.979108 |

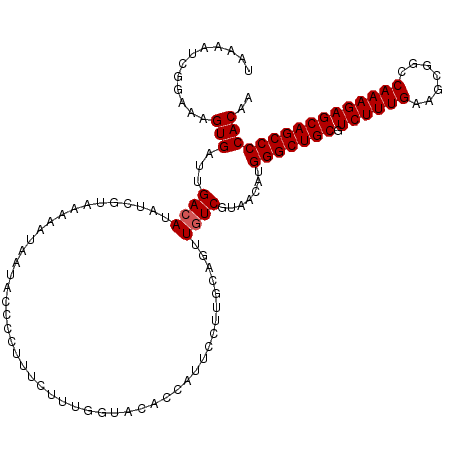
| Location | 15,836,015 – 15,836,119 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 87.65 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -27.95 |
| Energy contribution | -27.94 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15836015 104 + 23771897 UUGUGGGGCUGCUCUUUGGCCGCUUCAAAGACGCAGCCCAUGUUACGAAAACUGCA---------------AAGAAAGGGGUAUUAUUUUUACGAUAUGUCAAUCACUUUCCUAUCUUA (((((((((((((((((((.....))))))).)))))))....)))))........---------------((((.(((((((.......)))(((......)))....)))).)))). ( -30.50) >DroSec_CAF1 43109 119 + 1 UUGUGGGGCUGCUCUUUGGCCGCUUCAAAGACGCAGCCCAUGUUACGACAACUGGAAAGAAUGGUGUACCAAAGAAAGGGGUAUUAUUUUUACGAUAUGUCAAACACUUUCCAAUUUUA ..(((((((((((((((((.....))))))).))))))).......((((..(.(.((((((((((..((.......))..)))))))))).).)..))))...)))............ ( -38.40) >DroSim_CAF1 44290 119 + 1 UUGUGGGGCUGCUCUUUGGCCGCUUCAAAGACGCAGCCCAUGUUACGACAACUGCAAAGAAUGGUGUACCAAAGAAAGGGGUAUUAUUUUUACGAUAUGUCAAUCACUCUCCAAUUUUA ..(((((((((((((((((.....))))))).))))))).......((((......((((((((((..((.......))..))))))))))......))))...)))............ ( -37.10) >DroEre_CAF1 42133 119 + 1 UUGUGGGGCUGCUCUUUGGCCGCUUCAAAGACGCAGCCCAUGUUACGACAACUGCAAGGAGUGGUGAAUCAAACAGAAGGGUAUUAUUUUUAUGAUAUGUCAAUCACUUUCGGGUGUUA .....((((((((((((((.....))))))).))))))).......((((.(((...((((((((((.((.....))...(((((((....))))))).)).))))))))))).)))). ( -36.50) >DroYak_CAF1 84427 119 + 1 UUGUGGGGCUGCUCUUUGGCCGCUUCAAAGACGCAGCCCAUGUUACGACAACUGCAAGGAAUGGUUUGCCAAACAAAAGGGUAUUAUUUUUAUGACAUGUCAAUCACUUUCUGAUUUUA ..(((((((((((((((((.....))))))).))))))).......((((..(((((((((((((..(((.........)))))))))))).)).))))))...)))............ ( -34.50) >consensus UUGUGGGGCUGCUCUUUGGCCGCUUCAAAGACGCAGCCCAUGUUACGACAACUGCAAAGAAUGGUGUACCAAAGAAAGGGGUAUUAUUUUUACGAUAUGUCAAUCACUUUCCAAUUUUA ..(((((((((((((((((.....))))))).))))))).......((((...........(((....))).........(((.......)))....))))...)))............ (-27.95 = -27.94 + -0.01)

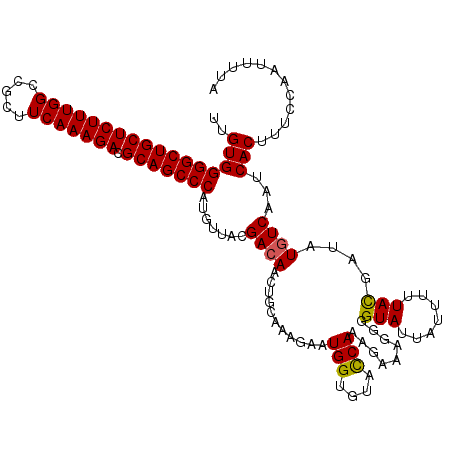

| Location | 15,836,015 – 15,836,119 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 87.65 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -23.87 |
| Energy contribution | -24.07 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15836015 104 - 23771897 UAAGAUAGGAAAGUGAUUGACAUAUCGUAAAAAUAAUACCCCUUUCUU---------------UGCAGUUUUCGUAACAUGGGCUGCGUCUUUGAAGCGGCCAAAGAGCAGCCCCACAA .((((.(((.....(((......)))(((.......))).))).))))---------------....(((.....)))..(((((((.((((((.......)))))))))))))..... ( -26.40) >DroSec_CAF1 43109 119 - 1 UAAAAUUGGAAAGUGUUUGACAUAUCGUAAAAAUAAUACCCCUUUCUUUGGUACACCAUUCUUUCCAGUUGUCGUAACAUGGGCUGCGUCUUUGAAGCGGCCAAAGAGCAGCCCCACAA ...(((((((((((((((.((.....))..))))).............(((....)))..))))))))))..........(((((((.((((((.......)))))))))))))..... ( -33.50) >DroSim_CAF1 44290 119 - 1 UAAAAUUGGAGAGUGAUUGACAUAUCGUAAAAAUAAUACCCCUUUCUUUGGUACACCAUUCUUUGCAGUUGUCGUAACAUGGGCUGCGUCUUUGAAGCGGCCAAAGAGCAGCCCCACAA ............(((...((((.((.((((((((..((((.........))))....))).))))).)))))).......(((((((.((((((.......)))))))))))))))).. ( -30.70) >DroEre_CAF1 42133 119 - 1 UAACACCCGAAAGUGAUUGACAUAUCAUAAAAAUAAUACCCUUCUGUUUGAUUCACCACUCCUUGCAGUUGUCGUAACAUGGGCUGCGUCUUUGAAGCGGCCAAAGAGCAGCCCCACAA ............(((...((((.....................((((..((........))...)))).)))).......(((((((.((((((.......)))))))))))))))).. ( -27.95) >DroYak_CAF1 84427 119 - 1 UAAAAUCAGAAAGUGAUUGACAUGUCAUAAAAAUAAUACCCUUUUGUUUGGCAAACCAUUCCUUGCAGUUGUCGUAACAUGGGCUGCGUCUUUGAAGCGGCCAAAGAGCAGCCCCACAA ............(((...((((........((((((.......)))))).((((........))))...)))).......(((((((.((((((.......)))))))))))))))).. ( -31.60) >consensus UAAAAUCGGAAAGUGAUUGACAUAUCGUAAAAAUAAUACCCCUUUCUUUGGUACACCAUUCCUUGCAGUUGUCGUAACAUGGGCUGCGUCUUUGAAGCGGCCAAAGAGCAGCCCCACAA ............(((...((((...............................................)))).......(((((((.((((((.......)))))))))))))))).. (-23.87 = -24.07 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:03:02 2006