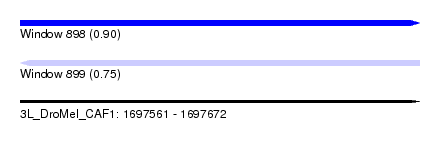
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,697,561 – 1,697,672 |
| Length | 111 |
| Max. P | 0.903967 |

| Location | 1,697,561 – 1,697,672 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.48 |
| Mean single sequence MFE | -47.12 |
| Consensus MFE | -16.83 |
| Energy contribution | -16.78 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.36 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1697561 111 + 23771897 UGCCUGCUGCCUGCCCCUUGGGAUUCUGCCCC-AGCAGG-CAAG--AG--CGGG--CUCCAGGAACACCCCCUGCUGGGAGUCCAAUGCGGAUGUCUUCCCCCGCCCAUGUGGCAACAU ((((.(((((((((....((((.......)))-))))))-)...--))--)(((--(..((((.......))))..((((((((.....))))....))))..))))....)))).... ( -44.60) >DroVir_CAF1 74919 111 + 1 UGCCAGCUGCCUGCCCCUGGGCAUAUUGCCCA---CGGG-UGGG--GGCAGCAG--CUCCAAGCCGACGCCCUGCUGGGAGUCCAAUGCGGACAUCUUUCCGCGACCCUGCGGCAACAU ((((.(((((((.(((((((((.....)))))---.)))-.).)--)))))).(--(........(((.(((....))).)))...((((((......)))))).....)))))).... ( -57.50) >DroPse_CAF1 66630 111 + 1 UGCCUGCUGCCUGCCGCUGGGAAUCCUCCCGC-AGGCGG-CUCG--AA--GCGG--CUCCCGGCACACUCCAUGCUGGGAAUCCAACGCCGACGUCUUUCCCCGUCCCUGCGGCAAUAU ((((.(((((((((....((((....))))))-))))))-)...--..--((((--(((((((((.......)))))))).......)))((((........))))...)))))).... ( -50.41) >DroWil_CAF1 72829 114 + 1 UGCCUGCUGCCUGCCACUGGGUAUACUCCCACCAGCUGGACGUG-GGU--CCAG--CUCUAGACAUACUCCCUGCUGGGAGUCCAAUGCCGAUGUUUUUCCGCGUCCAUGUGGCAAUAU ...........((((((((((......)))).....((((((((-((.--..((--(....(.(((((((((....))))))...))).)...)))..)))))))))).)))))).... ( -43.80) >DroAna_CAF1 57762 114 + 1 CACCUGCUGCUUGCCUUUGGGUAUCCUGCCUC-AACAGG-CUAGG-AA--UGGGUCCUCCAGAAACACCCCCUGUUGGGAGUCCAAUGCAGACGUCUUUCCCCGACCUUGCGGCAACAU ....((((((..(...(((((..((((((((.-...)))-).)))-).--((((..((((...((((.....)))).))))))))...............))))).)..)))))).... ( -36.00) >DroPer_CAF1 66779 111 + 1 UGCCUGCUGCCUGCCGCUGGGAAUCCUCCCGC-AGGCGG-CUCG--AA--GCGG--CUCCCGGCACACUCCAUGCUGGGAAUCCAACGCCGACGUCUUUCCCCGUCCCUGCGGCAAUAU ((((.(((((((((....((((....))))))-))))))-)...--..--((((--(((((((((.......)))))))).......)))((((........))))...)))))).... ( -50.41) >consensus UGCCUGCUGCCUGCCCCUGGGAAUCCUCCCCC_AGCAGG_CUAG__AA__GCGG__CUCCAGGCACACUCCCUGCUGGGAGUCCAAUGCCGACGUCUUUCCCCGUCCCUGCGGCAACAU ...........((((.(.((((......(((.....))).................((((.((((.......)))).)))).......................)))).).)))).... (-16.83 = -16.78 + -0.05)

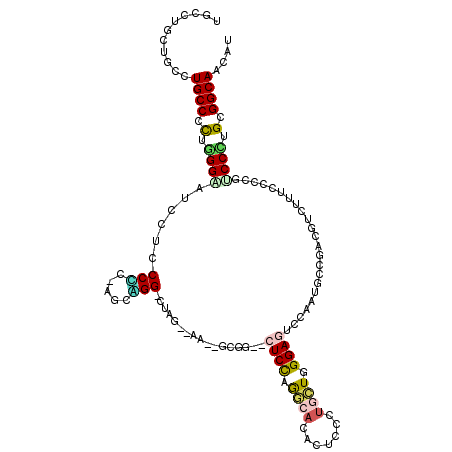
| Location | 1,697,561 – 1,697,672 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.48 |
| Mean single sequence MFE | -50.70 |
| Consensus MFE | -21.76 |
| Energy contribution | -20.43 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
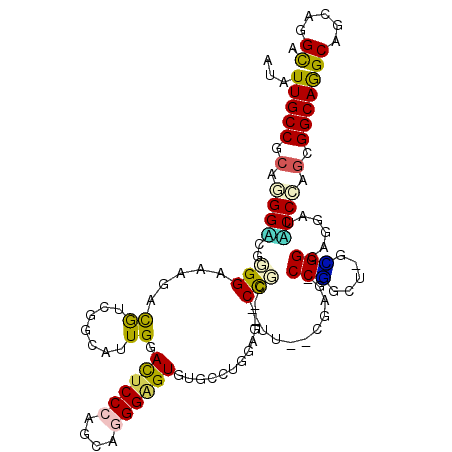
>3L_DroMel_CAF1 1697561 111 - 23771897 AUGUUGCCACAUGGGCGGGGGAAGACAUCCGCAUUGGACUCCCAGCAGGGGGUGUUCCUGGAG--CCCG--CU--CUUG-CCUGCU-GGGGCAGAAUCCCAAGGGGCAGGCAGCAGGCA ....((((....((((..(((((.((..((((..(((....))))).))..)).)))))...)--)))(--((--((((-(((..(-((((.....)))))..))))))).))).)))) ( -53.20) >DroVir_CAF1 74919 111 - 1 AUGUUGCCGCAGGGUCGCGGAAAGAUGUCCGCAUUGGACUCCCAGCAGGGCGUCGGCUUGGAG--CUGCUGCC--CCCA-CCCG---UGGGCAAUAUGCCCAGGGGCAGGCAGCUGGCA ....((((.(..(((((((((......)))))....(((.(((....))).)))))))..)((--((((((((--((..-....---(((((.....))))))))))).)))))))))) ( -58.40) >DroPse_CAF1 66630 111 - 1 AUAUUGCCGCAGGGACGGGGAAAGACGUCGGCGUUGGAUUCCCAGCAUGGAGUGUGCCGGGAG--CCGC--UU--CGAG-CCGCCU-GCGGGAGGAUUCCCAGCGGCAGGCAGCAGGCA ...((((((((((((((........))))((((((((....))))).(((((((.((.....)--))))--))--)).)-)).)))-))(((......)))...)))))((.....)). ( -50.40) >DroWil_CAF1 72829 114 - 1 AUAUUGCCACAUGGACGCGGAAAAACAUCGGCAUUGGACUCCCAGCAGGGAGUAUGUCUAGAG--CUGG--ACC-CACGUCCAGCUGGUGGGAGUAUACCCAGUGGCAGGCAGCAGGCA ...(((((((.(((((((.((......)).)).....((((((....))))))..))))).((--((((--((.-...))))))))..((((......)))))))))))((.....)). ( -49.80) >DroAna_CAF1 57762 114 - 1 AUGUUGCCGCAAGGUCGGGGAAAGACGUCUGCAUUGGACUCCCAACAGGGGGUGUUUCUGGAGGACCCA--UU-CCUAG-CCUGUU-GAGGCAGGAUACCCAAAGGCAAGCAGCAGGUG .(((((((....)(((.(((......((((.((..(.((((((....)))))).)...))..))))...--.(-(((.(-(((...-.))))))))..)))...)))..)))))).... ( -42.00) >DroPer_CAF1 66779 111 - 1 AUAUUGCCGCAGGGACGGGGAAAGACGUCGGCGUUGGAUUCCCAGCAUGGAGUGUGCCGGGAG--CCGC--UU--CGAG-CCGCCU-GCGGGAGGAUUCCCAGCGGCAGGCAGCAGGCA ...((((((((((((((........))))((((((((....))))).(((((((.((.....)--))))--))--)).)-)).)))-))(((......)))...)))))((.....)). ( -50.40) >consensus AUAUUGCCGCAGGGACGGGGAAAGACGUCGGCAUUGGACUCCCAGCAGGGAGUGUGCCUGGAG__CCGC__UU__CGAG_CCGGCU_GCGGCAGGAUACCCAGCGGCAGGCAGCAGGCA ...(((((.(.((((..(((.....((.......)).((((((....))))))............)))............(((.....)))......)))).).)))))((.....)). (-21.76 = -20.43 + -1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:46:02 2006