
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
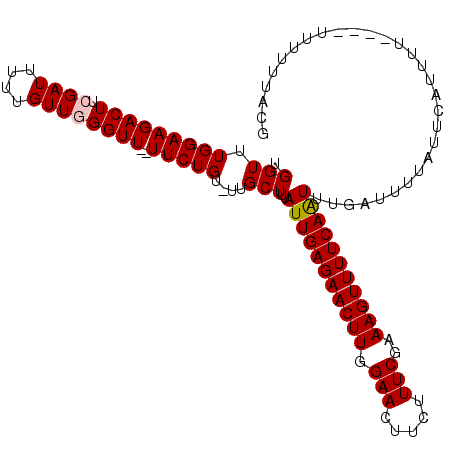
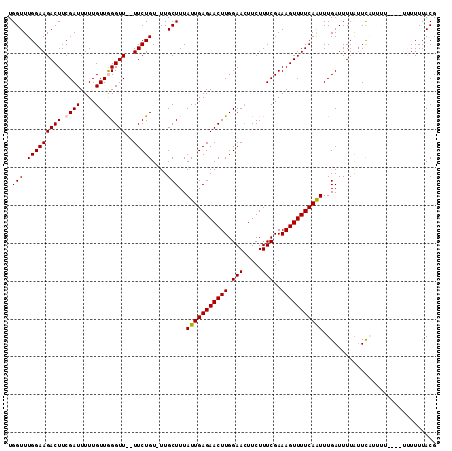
| Location | 15,828,235 – 15,828,339 |
| Length | 104 |
| Max. P | 0.599772 |

| Location | 15,828,235 – 15,828,339 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 90.15 |
| Mean single sequence MFE | -19.74 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.46 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15828235 104 + 23771897 UGGUUUGGAAGACUUCGAUUUUUGUUGGGUU--UUCUGU-UUGCUUUAUUGAGAACUUGGAACUUCUUUCGAAAGUUUUCAAUUUGAUUUUAGUCAUUUU------UUUUACG .(((.(((((((((.((((....))))))))--))))).-..)))..(((((((((((.(((.....)))..))))))))))).((((....))))....------....... ( -21.80) >DroSec_CAF1 35583 113 + 1 UGGUUUGGAAGACUUCGAUUUUUGUUGGGUUUUUUCUGUUUUGCUUUAUUGAGAACUUGGAAAUUCUUUCGAAAGUUUUCAAUUUGAUUUUAUUCAUUUUUUUAUUUUUUACG .......(((((((((((......(..(((((((..((........))..)))))))..)........))).))))))))....(((......)))................. ( -18.74) >DroSim_CAF1 36817 111 + 1 UGGUUUGGAAGACUUCGAUUUUUGUUGGGUUU-UUCUGU-UUGCUUUAUUGAGAACUUGGAACUUCUUUCGAAAGUUUUCAAUUUGAUUUUAUUCAUUUUUUUAUUUUUUACG .(((.(((((((.((((((....)))))).))-))))).-..)))..(((((((((((.(((.....)))..))))))))))).(((......)))................. ( -19.40) >DroEre_CAF1 34792 100 + 1 UGGUUUGGAAGACUUCGAUUUUUGUUAGGUU--UUCUGU-UUGCUUUAUUGAGAACUUGGAACUUCUUUCGAAAGUUUUCAGUUUGAUUUUGUUU----G------UUUUACG .......(((((((((((......(((((((--(((...-..........))))))))))........))).)))))))).((..(((.......----)------))..)). ( -19.46) >DroYak_CAF1 77097 102 + 1 UGGUUUGGAAGACUUCGAUUUUUGUUAGGUU--UUCUGU-UUGCUUUAUUGAGAACUUGGAACUUCUUUCGAAAGUUUUCAAUUUGAUUUUGUUU----U----UUUUUUACG .(((.((((((((((.(((....))))))))--))))).-..)))..(((((((((((.(((.....)))..)))))))))))............----.----......... ( -19.30) >consensus UGGUUUGGAAGACUUCGAUUUUUGUUGGGUU__UUCUGU_UUGCUUUAUUGAGAACUUGGAACUUCUUUCGAAAGUUUUCAAUUUGAUUUUAUUCAUUUU____UUUUUUACG .(((.(((((((((.((((....))))))))..)))))....)))..(((((((((((.(((.....)))..))))))))))).............................. (-14.22 = -14.46 + 0.24)

| Location | 15,828,235 – 15,828,339 |
|---|---|
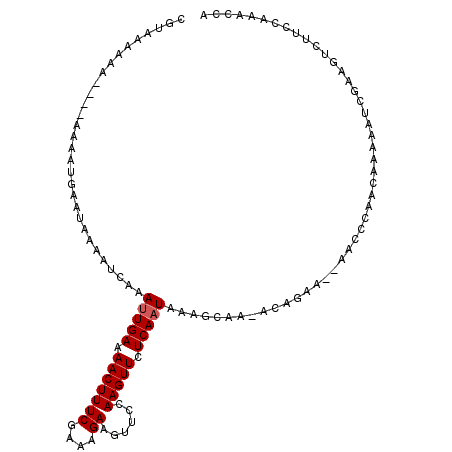
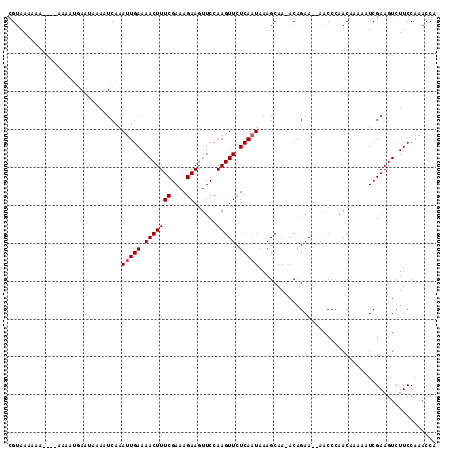
| Length | 104 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 90.15 |
| Mean single sequence MFE | -8.34 |
| Consensus MFE | -7.40 |
| Energy contribution | -7.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15828235 104 - 23771897 CGUAAAA------AAAAUGACUAAAAUCAAAUUGAAAACUUUCGAAAGAAGUUCCAAGUUCUCAAUAAAGCAA-ACAGAA--AACCCAACAAAAAUCGAAGUCUUCCAAACCA .......------.....((((........(((((.(((((((....))......))))).))))).......-...(..--....)............)))).......... ( -9.80) >DroSec_CAF1 35583 113 - 1 CGUAAAAAAUAAAAAAAUGAAUAAAAUCAAAUUGAAAACUUUCGAAAGAAUUUCCAAGUUCUCAAUAAAGCAAAACAGAAAAAACCCAACAAAAAUCGAAGUCUUCCAAACCA .((..............(((......))).(((((.(((((((....))......))))).)))))...)).......................................... ( -8.40) >DroSim_CAF1 36817 111 - 1 CGUAAAAAAUAAAAAAAUGAAUAAAAUCAAAUUGAAAACUUUCGAAAGAAGUUCCAAGUUCUCAAUAAAGCAA-ACAGAA-AAACCCAACAAAAAUCGAAGUCUUCCAAACCA .((..............(((......))).(((((.(((((((....))......))))).)))))...))..-......-................................ ( -8.40) >DroEre_CAF1 34792 100 - 1 CGUAAAA------C----AAACAAAAUCAAACUGAAAACUUUCGAAAGAAGUUCCAAGUUCUCAAUAAAGCAA-ACAGAA--AACCUAACAAAAAUCGAAGUCUUCCAAACCA .......------.----...........((((...(((((.(....))))))...)))).............-......--............................... ( -7.10) >DroYak_CAF1 77097 102 - 1 CGUAAAAAA----A----AAACAAAAUCAAAUUGAAAACUUUCGAAAGAAGUUCCAAGUUCUCAAUAAAGCAA-ACAGAA--AACCUAACAAAAAUCGAAGUCUUCCAAACCA .........----.----............(((((.(((((((....))......))))).))))).......-......--............................... ( -8.00) >consensus CGUAAAAAA____AAAAUGAAUAAAAUCAAAUUGAAAACUUUCGAAAGAAGUUCCAAGUUCUCAAUAAAGCAA_ACAGAA__AACCCAACAAAAAUCGAAGUCUUCCAAACCA ..............................(((((.(((((((....))......))))).)))))............................................... ( -7.40 = -7.60 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:53 2006