| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,827,247 – 15,827,337 |
| Length | 90 |
| Max. P | 0.932489 |

| Location | 15,827,247 – 15,827,337 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.80 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -22.85 |
| Energy contribution | -23.72 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
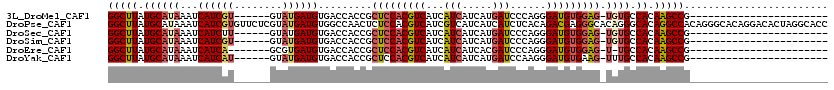
>3L_DroMel_CAF1 15827247 90 + 23771897 GGCUUAUGCAUAAAUCAUCGU------GUAUGAUGUGACCACCGCUCCACGUCAUCAUCAUCAUGAUCCCAGGGAUGUGGAG-UGUGCCACAAGCCG----------------------- (((((((((((........))------))))))((((..(((.(((((((((((((((....)))))(....))))))))))-)))).)))).))).----------------------- ( -33.90) >DroPse_CAF1 33917 120 + 1 GGCUUAUGCAUAAAUCAUCGUGUUCUCGUAUGAUGUGGCCAACUCUCCACGUCAUCGUCAUCAUCAUCUCACAGACGAGGGCACAGGGCACAGGCCACAGGGCACAGGACACUAGGCACC (((((.(((..........(((((((((.(((((((((........))))))))))(((..............)))))))))))...))).)))))...((((..((....))..)).)) ( -37.96) >DroSec_CAF1 34615 90 + 1 GGCUUAUGCAUAAAUCAUCUU------GUAUGAUGUGACCACCGCUCCACGUCAUCAUCAUCAUGAUCCCAGGGAUGUGGAG-UGUGCCACAAGCCG----------------------- ((((((((((..........)------))))))((((..(((.(((((((((((((((....)))))(....))))))))))-)))).)))).))).----------------------- ( -32.40) >DroSim_CAF1 35813 90 + 1 GGCUUAUGCAUAAAUCAUCGU------GUAUGAUGUGACCACCGCUCCACGUCAUCAUCAUCAUGAUCCCAGGGAUGUGGAG-UGUGCCACAAGCCG----------------------- (((((((((((........))------))))))((((..(((.(((((((((((((((....)))))(....))))))))))-)))).)))).))).----------------------- ( -33.90) >DroEre_CAF1 33542 88 + 1 GGCUUAUGCAUAAAUCAUCA-------GCGUGAUGUGACCACCGCUCCACGUCAUCAUCAUCACGAUCCCAGGGAUGUGGAG-U-UGCCACAAGCCG----------------------- (((((..(((.....((((.-------....))))........((((((((((...(((.....)))......)))))))))-)-)))...))))).----------------------- ( -27.70) >DroYak_CAF1 75848 90 + 1 GGCUUAUGCAUAAAUCAUCAU------GUAUGAUGUGACCACCGCUCCACGUCAUCAUCAUCAUGAUCCAAGGGAUGUGAAG-UUUGCCACAAGCCG----------------------- (((((..(((.(..(((((((------(.((((((((((...........)))).))))))))))((((...))))))))..-).)))...))))).----------------------- ( -27.00) >consensus GGCUUAUGCAUAAAUCAUCGU______GUAUGAUGUGACCACCGCUCCACGUCAUCAUCAUCAUGAUCCCAGGGAUGUGGAG_UGUGCCACAAGCCG_______________________ (((((.((((((...((((((........)))))).........(((((((((...(((.....)))......))))))))).)))).)).)))))........................ (-22.85 = -23.72 + 0.86)


| Location | 15,827,247 – 15,827,337 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.80 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -23.47 |
| Energy contribution | -24.08 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15827247 90 - 23771897 -----------------------CGGCUUGUGGCACA-CUCCACAUCCCUGGGAUCAUGAUGAUGAUGACGUGGAGCGGUGGUCACAUCAUAC------ACGAUGAUUUAUGCAUAAGCC -----------------------.((((((((.((..-..((((.(((.((..(((((....)))))..)).)))...))))...((((....------..)))).....)))))))))) ( -30.60) >DroPse_CAF1 33917 120 - 1 GGUGCCUAGUGUCCUGUGCCCUGUGGCCUGUGCCCUGUGCCCUCGUCUGUGAGAUGAUGAUGACGAUGACGUGGAGAGUUGGCCACAUCAUACGAGAACACGAUGAUUUAUGCAUAAGCC .((((...((((.(((((...(((((((...((((..((.(.(((((..............))))).).))..).).)).)))))))...))).)).))))((....))..))))..... ( -31.94) >DroSec_CAF1 34615 90 - 1 -----------------------CGGCUUGUGGCACA-CUCCACAUCCCUGGGAUCAUGAUGAUGAUGACGUGGAGCGGUGGUCACAUCAUAC------AAGAUGAUUUAUGCAUAAGCC -----------------------.((((((((.((..-..((((.(((.((..(((((....)))))..)).)))...))))...((((....------..)))).....)))))))))) ( -30.60) >DroSim_CAF1 35813 90 - 1 -----------------------CGGCUUGUGGCACA-CUCCACAUCCCUGGGAUCAUGAUGAUGAUGACGUGGAGCGGUGGUCACAUCAUAC------ACGAUGAUUUAUGCAUAAGCC -----------------------.((((((((.((..-..((((.(((.((..(((((....)))))..)).)))...))))...((((....------..)))).....)))))))))) ( -30.60) >DroEre_CAF1 33542 88 - 1 -----------------------CGGCUUGUGGCA-A-CUCCACAUCCCUGGGAUCGUGAUGAUGAUGACGUGGAGCGGUGGUCACAUCACGC-------UGAUGAUUUAUGCAUAAGCC -----------------------.((((((((.((-.-..((((.(((.((..(((((....)))))..)).)))...))))...((((....-------.)))).....)))))))))) ( -30.60) >DroYak_CAF1 75848 90 - 1 -----------------------CGGCUUGUGGCAAA-CUUCACAUCCCUUGGAUCAUGAUGAUGAUGACGUGGAGCGGUGGUCACAUCAUAC------AUGAUGAUUUAUGCAUAAGCC -----------------------.((((((((.((..-..(((..(((...)))(((((((((((.((((...........)))))))))).)------)))))))....)))))))))) ( -29.40) >consensus _______________________CGGCUUGUGGCACA_CUCCACAUCCCUGGGAUCAUGAUGAUGAUGACGUGGAGCGGUGGUCACAUCAUAC______ACGAUGAUUUAUGCAUAAGCC ........................((((((((........((((.(((.((..(((((....)))))..)).)))...))))...((((............)))).......)))))))) (-23.47 = -24.08 + 0.61)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:49 2006