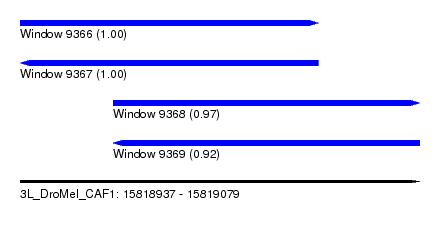
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,818,937 – 15,819,079 |
| Length | 142 |
| Max. P | 0.999967 |

| Location | 15,818,937 – 15,819,043 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -44.80 |
| Consensus MFE | -40.98 |
| Energy contribution | -41.62 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.84 |
| SVM RNA-class probability | 0.999955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15818937 106 + 23771897 GGCUGACUUGUCUUUGUUUGCCAGCAGCUGGCGGCCAUUUGCCGAGUGCACAAAUGCCGGCAAAGGUGGCCGGUGG-GGUUGCGAAAAUGGAAAAGUCCG-GGAAAAU ..(((((((.(((...(((((.(((.((((((.(((.(((((((.((........)))))))))))).))))))..-.))))))))...))).)))).))-)...... ( -45.10) >DroSec_CAF1 26332 107 + 1 GGCUGACUUGUCUUUGUUUGCCAGCAGCUGGCGGCCAUUUGCCGAGUGCACAAAUGCCGGCAAAGGUGGCCGGUGG-GGUUGCGAAAAUGGAAAAGUCUGGGGAAAAU ....(((((.(((...(((((.(((.((((((.(((.(((((((.((........)))))))))))).))))))..-.))))))))...))).))))).......... ( -45.10) >DroSim_CAF1 26917 107 + 1 GGCUGACUUGUCUUUGUUUGCCAUCAGCUGGCGGCCAUUUGCCGAGUGCACAAAUGCCGGCAAAGGUGGCCGGUGG-GGUUGCGAAAAUGGAAAAGUCUGGGGAAAAU ....(((((.(((...(((((.(((.((((((.(((.(((((((.((........)))))))))))).))))))..-))).)))))...))).))))).......... ( -45.50) >DroEre_CAF1 25268 107 + 1 GGCUGACUUGACUUUGUUUGCCAGCAGCUGGCGGCCAUUUGCCGAGUGCACAAAUGCCGGCAAAGGUGGCCGGUGGGGGUUGCGAAAAUGGAAAAGUGGG-UGGAAAU .(((.((((..((...(((((.(((.((((((.(((.(((((((.((........)))))))))))).))))))....))))))))...))..)))).))-)...... ( -44.70) >DroYak_CAF1 67499 106 + 1 GGCUGACUUGUCUUUGUUUGCCAGCAGCUGGCGGCCAUUUGCCGAGUGCACAAAUGCCGGCAAAGGUGGCCGGUGG-GGUUGCGAGAAUGGAAAAGUGGU-GGAAAAC .(((.((((.(((...(((((.(((.((((((.(((.(((((((.((........)))))))))))).))))))..-.))))))))...))).)))))))-....... ( -43.60) >consensus GGCUGACUUGUCUUUGUUUGCCAGCAGCUGGCGGCCAUUUGCCGAGUGCACAAAUGCCGGCAAAGGUGGCCGGUGG_GGUUGCGAAAAUGGAAAAGUCGG_GGAAAAU ....(((((.(((...(((((.(((.((((((.(((.(((((((.((........)))))))))))).))))))....))))))))...))).))))).......... (-40.98 = -41.62 + 0.64)



| Location | 15,818,937 – 15,819,043 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 95.14 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -30.64 |
| Energy contribution | -31.44 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.95 |
| SVM decision value | 5.00 |
| SVM RNA-class probability | 0.999967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15818937 106 - 23771897 AUUUUCC-CGGACUUUUCCAUUUUCGCAACC-CCACCGGCCACCUUUGCCGGCAUUUGUGCACUCGGCAAAUGGCCGCCAGCUGCUGGCAAACAAAGACAAGUCAGCC .......-..(((((.((.............-.....(((((..((((((((((....)))...))))))))))))(((((...))))).......)).))))).... ( -34.20) >DroSec_CAF1 26332 107 - 1 AUUUUCCCCAGACUUUUCCAUUUUCGCAACC-CCACCGGCCACCUUUGCCGGCAUUUGUGCACUCGGCAAAUGGCCGCCAGCUGCUGGCAAACAAAGACAAGUCAGCC ..........(((((.((.............-.....(((((..((((((((((....)))...))))))))))))(((((...))))).......)).))))).... ( -34.40) >DroSim_CAF1 26917 107 - 1 AUUUUCCCCAGACUUUUCCAUUUUCGCAACC-CCACCGGCCACCUUUGCCGGCAUUUGUGCACUCGGCAAAUGGCCGCCAGCUGAUGGCAAACAAAGACAAGUCAGCC ..........(((((.((.............-.....(((((..((((((((((....)))...))))))))))))((((.....)))).......)).))))).... ( -33.50) >DroEre_CAF1 25268 107 - 1 AUUUCCA-CCCACUUUUCCAUUUUCGCAACCCCCACCGGCCACCUUUGCCGGCAUUUGUGCACUCGGCAAAUGGCCGCCAGCUGCUGGCAAACAAAGUCAAGUCAGCC .......-............................((((((..((((((((((....)))...)))))))))))))...(((((((((.......))).)).)))). ( -29.90) >DroYak_CAF1 67499 106 - 1 GUUUUCC-ACCACUUUUCCAUUCUCGCAACC-CCACCGGCCACCUUUGCCGGCAUUUGUGCACUCGGCAAAUGGCCGCCAGCUGCUGGCAAACAAAGACAAGUCAGCC .......-...((((.((.............-.....(((((..((((((((((....)))...))))))))))))(((((...))))).......)).))))..... ( -30.00) >consensus AUUUUCC_CAGACUUUUCCAUUUUCGCAACC_CCACCGGCCACCUUUGCCGGCAUUUGUGCACUCGGCAAAUGGCCGCCAGCUGCUGGCAAACAAAGACAAGUCAGCC ..........(((((.((...................(((((..((((((((((....)))...))))))))))))(((((...))))).......)).))))).... (-30.64 = -31.44 + 0.80)

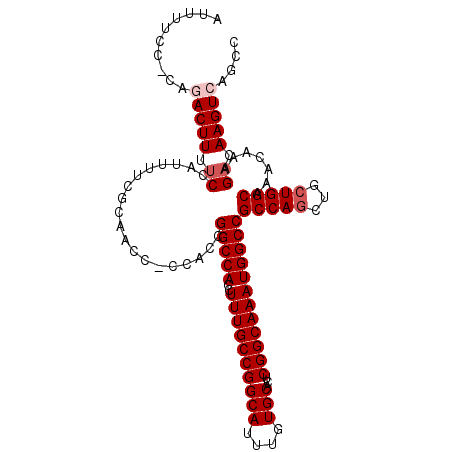
| Location | 15,818,970 – 15,819,079 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 91.46 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -24.14 |
| Energy contribution | -24.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15818970 109 + 23771897 GCCAUUUGCCGAGUGCACAAAUGCCGGCAAAGGUGGCCGGUGG-GGUUGCGAAAAUGGAAAAGUCCG-GGAAAAUGGGUAAGUGGGAAAGUGGGCUGGGAGGAAACAGUUA .(((((((((...(((((...(((((((.......))))))).-.).))))....((((....))))-........))))))))).......(((((........))))). ( -35.80) >DroSec_CAF1 26365 110 + 1 GCCAUUUGCCGAGUGCACAAAUGCCGGCAAAGGUGGCCGGUGG-GGUUGCGAAAAUGGAAAAGUCUGGGGAAAAUGGGAAAGUGGAAAAGUGGGCGGCGAGGAAACAGUUA (((.(((.(((..(((((...(((((((.......))))))).-.).))))............(((.(......).)))...))).)))...)))(((..(....).))). ( -26.80) >DroSim_CAF1 26950 110 + 1 GCCAUUUGCCGAGUGCACAAAUGCCGGCAAAGGUGGCCGGUGG-GGUUGCGAAAAUGGAAAAGUCUGGGGAAAAUGGGAAAGUGGAAAAGUGGGCGGGGAGGAAACAGUUA (((.(((.(((..(((((...(((((((.......))))))).-.).))))............(((.(......).)))...))).)))...))).....(....)..... ( -26.20) >DroEre_CAF1 25301 110 + 1 GCCAUUUGCCGAGUGCACAAAUGCCGGCAAAGGUGGCCGGUGGGGGUUGCGAAAAUGGAAAAGUGGG-UGGAAAUGGGAAAAUGGGAAAGUGGGCUGGCAGGAAGCAGUUA ((..((((((.(((.(((....((((((.......))))))...(.(..(............)..).-)....................))).)))))))))..))..... ( -29.80) >DroYak_CAF1 67532 109 + 1 GCCAUUUGCCGAGUGCACAAAUGCCGGCAAAGGUGGCCGGUGG-GGUUGCGAGAAUGGAAAAGUGGU-GGAAAACAGGAAAAAGGGGAAAUGGGCUGGGAGGAAACAGUUA .((((((.((...(((((...(((((((.......))))))).-.).))))...........((...-.....)).........)).))))))((((........)))).. ( -29.70) >consensus GCCAUUUGCCGAGUGCACAAAUGCCGGCAAAGGUGGCCGGUGG_GGUUGCGAAAAUGGAAAAGUCGG_GGAAAAUGGGAAAGUGGGAAAGUGGGCUGGGAGGAAACAGUUA (((.(((((((.((........))))))))).(..(((......)))..)..........................................))).....(....)..... (-24.14 = -24.14 + 0.00)



| Location | 15,818,970 – 15,819,079 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 91.46 |
| Mean single sequence MFE | -19.28 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.30 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15818970 109 - 23771897 UAACUGUUUCCUCCCAGCCCACUUUCCCACUUACCCAUUUUCC-CGGACUUUUCCAUUUUCGCAACC-CCACCGGCCACCUUUGCCGGCAUUUGUGCACUCGGCAAAUGGC ...(((........))).................((((((.((-.(((....)))......(((...-...(((((.......)))))......)))....)).)))))). ( -23.00) >DroSec_CAF1 26365 110 - 1 UAACUGUUUCCUCGCCGCCCACUUUUCCACUUUCCCAUUUUCCCCAGACUUUUCCAUUUUCGCAACC-CCACCGGCCACCUUUGCCGGCAUUUGUGCACUCGGCAAAUGGC ..................................((((((.((...((....)).......(((...-...(((((.......)))))......)))....)).)))))). ( -18.20) >DroSim_CAF1 26950 110 - 1 UAACUGUUUCCUCCCCGCCCACUUUUCCACUUUCCCAUUUUCCCCAGACUUUUCCAUUUUCGCAACC-CCACCGGCCACCUUUGCCGGCAUUUGUGCACUCGGCAAAUGGC ..................................((((((.((...((....)).......(((...-...(((((.......)))))......)))....)).)))))). ( -18.20) >DroEre_CAF1 25301 110 - 1 UAACUGCUUCCUGCCAGCCCACUUUCCCAUUUUCCCAUUUCCA-CCCACUUUUCCAUUUUCGCAACCCCCACCGGCCACCUUUGCCGGCAUUUGUGCACUCGGCAAAUGGC .....(((...((((((....))....................-.................(((.......(((((.......)))))......)))....))))...))) ( -18.42) >DroYak_CAF1 67532 109 - 1 UAACUGUUUCCUCCCAGCCCAUUUCCCCUUUUUCCUGUUUUCC-ACCACUUUUCCAUUCUCGCAACC-CCACCGGCCACCUUUGCCGGCAUUUGUGCACUCGGCAAAUGGC ...(((........))).((((((.((.........((.....-...))............(((...-...(((((.......)))))......)))....)).)))))). ( -18.60) >consensus UAACUGUUUCCUCCCAGCCCACUUUCCCACUUUCCCAUUUUCC_CAGACUUUUCCAUUUUCGCAACC_CCACCGGCCACCUUUGCCGGCAUUUGUGCACUCGGCAAAUGGC ..........................................................................((((..((((((((((....)))...))))))))))) (-15.30 = -15.30 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:42 2006