
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,811,464 – 15,811,592 |
| Length | 128 |
| Max. P | 0.949462 |

| Location | 15,811,464 – 15,811,559 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 79.74 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -15.16 |
| Energy contribution | -15.22 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15811464 95 + 23771897 GGGCUCGUAAUAUCCUUAUAAUGGCACCAGCAGCUCUCCGGGGGCAUUAUUACUACAAUACUACUAUUCCAGCACUUUUCU-UUUUAGUGUGGGAA ((((.(((.(((....))).))))).))....((((.....)))).....................((((.(((((.....-....))))).)))) ( -20.10) >DroSec_CAF1 18972 85 + 1 GGGCUCGUAAUAUCCUUAUAAUGGCACCAGCAGCUCUCCGGGGGCUUUAUUAC--------UACUAUUCCUGCACUU---UCUUUUAGUGUGGGAA ((((.(((.(((....))).))))).))...(((((.....))))).......--------.....((((..((((.---......))))..)))) ( -23.70) >DroSim_CAF1 19382 88 + 1 UGGCUCGUAAUAUCCUUAUAAUGGCACCCCCCCCCCUCCAGGGGCAUUAUUAC--------UACUAUUCCAGCACUUUUCUCUUUUAGUGUGGGAA ......((((((.((.......))........((((....))))...))))))--------.....((((.(((((..........))))).)))) ( -19.10) >DroYak_CAF1 57294 75 + 1 GGGCUCGUAAUAUCCUUAUAAUGGC-CUGG-----CACCAGGGGGAUUAUUGC--------UACUAUUCCCGCAU-------UUUUAGUGUGGGAA ......((((((((((........(-(((.-----...)))))))).))))))--------.....(((((((((-------.....))))))))) ( -25.10) >consensus GGGCUCGUAAUAUCCUUAUAAUGGCACCAGCAGCUCUCCAGGGGCAUUAUUAC________UACUAUUCCAGCACUU___U_UUUUAGUGUGGGAA ......((((((.((((....(((.............)))))))...)))))).............((((((((((..........)))))))))) (-15.16 = -15.22 + 0.06)



| Location | 15,811,495 – 15,811,592 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 82.69 |
| Mean single sequence MFE | -21.89 |
| Consensus MFE | -14.85 |
| Energy contribution | -15.21 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
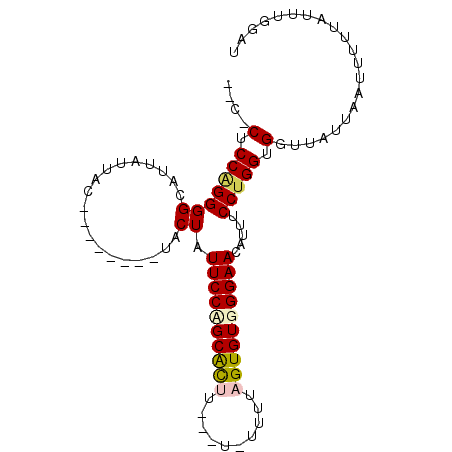
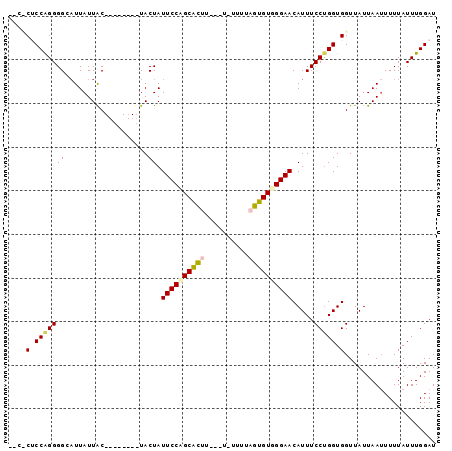
>3L_DroMel_CAF1 15811495 97 + 23771897 AGCUCUCCGGGGGCAUUAUUACUACAAUACUACUAUUCCAGCACUUUUCU-UUUUAGUGUGGGAACAUUUCCUGGUGGUUAUUAAAUUUUAUUUGGAU .((((.....))))...........((((((((((((((.(((((.....-....))))).)))).......)))))).))))............... ( -18.91) >DroSec_CAF1 19003 87 + 1 AGCUCUCCGGGGGCUUUAUUAC--------UACUAUUCCUGCACUU---UCUUUUAGUGUGGGAACAUUUCCUGGUGGUUAUUAAUUUUUAUUUGGAU (((((.....)))))(((..((--------(((((((((..((((.---......))))..)))).......)))))))...)))............. ( -22.41) >DroSim_CAF1 19413 90 + 1 CCCCCUCCAGGGGCAUUAUUAC--------UACUAUUCCAGCACUUUUCUCUUUUAGUGUGGGAACAUUUCCUGGUGGUUAUUAAUUUUUAUUUGGAU .((((....)))).((((..((--------(((((((((.(((((..........))))).)))).......)))))))...))))............ ( -21.31) >DroEre_CAF1 18316 79 + 1 ----CACCAGGGGGAUUAUUGC--------UACUAUUCCCGCGU-------UUUUAGUGUGGGAACAUUUCCCGGUGGUUAUUAAUUUUUAUUUGGAU ----((((.(((..((......--------.....((((((((.-------......)))))))).))..)))))))..................... ( -21.30) >DroYak_CAF1 57323 79 + 1 ----CACCAGGGGGAUUAUUGC--------UACUAUUCCCGCAU-------UUUUAGUGUGGGAACAUUUCCUGGUGGUUAUUAAUUUUUAUUUGGAU ----..(((((.((((((..((--------((((((((((((((-------.....))))))))).......)))))))...))))))...))))).. ( -25.51) >consensus __C_CUCCAGGGGCAUUAUUAC________UACUAUUCCAGCACUU___U_UUUUAGUGUGGGAACAUUUCCUGGUGGUUAUUAAUUUUUAUUUGGAU ....(.(((((((...................)).((((((((((..........)))))))))).....))))).)..................... (-14.85 = -15.21 + 0.36)

| Location | 15,811,495 – 15,811,592 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 82.69 |
| Mean single sequence MFE | -18.30 |
| Consensus MFE | -8.88 |
| Energy contribution | -10.20 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
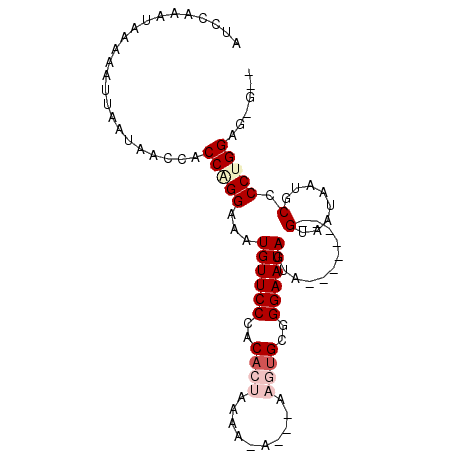
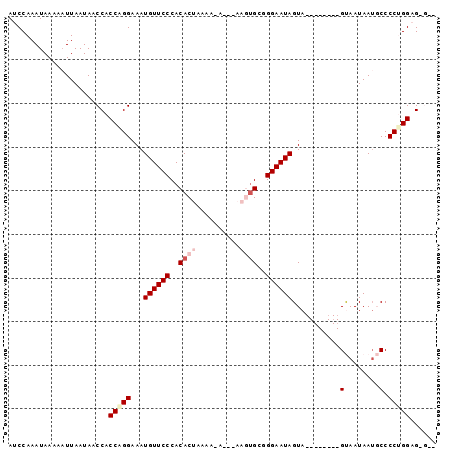
>3L_DroMel_CAF1 15811495 97 - 23771897 AUCCAAAUAAAAUUUAAUAACCACCAGGAAAUGUUCCCACACUAAAA-AGAAAAGUGCUGGAAUAGUAGUAUUGUAGUAAUAAUGCCCCCGGAGAGCU .......................((.((...((((((..((((....-.....))))..))))))...(((((((....))))))).)).))...... ( -17.30) >DroSec_CAF1 19003 87 - 1 AUCCAAAUAAAAAUUAAUAACCACCAGGAAAUGUUCCCACACUAAAAGA---AAGUGCAGGAAUAGUA--------GUAAUAAAGCCCCCGGAGAGCU .(((................((....))...((((((..((((......---.))))..))))))...--------..............)))..... ( -14.10) >DroSim_CAF1 19413 90 - 1 AUCCAAAUAAAAAUUAAUAACCACCAGGAAAUGUUCCCACACUAAAAGAGAAAAGUGCUGGAAUAGUA--------GUAAUAAUGCCCCUGGAGGGGG ....................((.(((((...((((((..((((..........))))..))))))(((--------.......))).))))).))... ( -20.80) >DroEre_CAF1 18316 79 - 1 AUCCAAAUAAAAAUUAAUAACCACCGGGAAAUGUUCCCACACUAAAA-------ACGCGGGAAUAGUA--------GCAAUAAUCCCCCUGGUG---- .....................(((((((...(((((((.(.......-------..).))))))).((--------....)).....)))))))---- ( -18.70) >DroYak_CAF1 57323 79 - 1 AUCCAAAUAAAAAUUAAUAACCACCAGGAAAUGUUCCCACACUAAAA-------AUGCGGGAAUAGUA--------GCAAUAAUCCCCCUGGUG---- .....................(((((((...(((((((.((......-------.)).))))))).((--------....)).....)))))))---- ( -20.60) >consensus AUCCAAAUAAAAAUUAAUAACCACCAGGAAAUGUUCCCACACUAAAA_A___AAGUGCGGGAAUAGUA________GUAAUAAUGCCCCUGGAG_G__ .......................(((((...((((((..((((..........))))..))))))...........(........).)))))...... ( -8.88 = -10.20 + 1.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:31 2006