| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,810,338 – 15,810,447 |
| Length | 109 |
| Max. P | 0.683366 |

| Location | 15,810,338 – 15,810,447 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.80 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -16.23 |
| Energy contribution | -16.95 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15810338 109 - 23771897 AAAAAAAAAUUGAAUCGAAAAGCAAAUGCUACCCAUUAACAAUAAAAUACACA-GCGAAAAAACAGUGGGGGUGGCAAAUAGGCUGGUCGAAGAGUGGGCGAAAUGGAUU---------- ..............((((..(((...((((((((.(((....)))....(((.-...........))).)))))))).....)))..))))...................---------- ( -22.80) >DroPse_CAF1 15435 109 - 1 CAAC-AAAAUCGAAUCGAAAAGCAAAUGCUAACCAUUAACAAUAAAAUACAAAAGCCAAA-UGUA-------UGGCGGCUC--CUGGGGGGCGGGUGUGGGUUGUGGGUUGUGUGUGUUG ....-...............((((.((((.((((....(((((...((((....((((..-....-------)))).((((--(...)))))..))))..))))).)))))))).)))). ( -29.20) >DroSec_CAF1 17897 108 - 1 GAAA-AAAAUGGAAUCGAAAAGCAAAUGCUACCCAUUAACAAUAAAAUACACA-GCGAAAAAACAGUGGGGGUGGCAAAUAGGCUGGUCGAAGAGUGGGCGAAAUGGUUU---------- ....-.........((((..(((...((((((((.(((....)))....(((.-...........))).)))))))).....)))..))))...................---------- ( -22.80) >DroSim_CAF1 18214 108 - 1 GAAA-AAAAUGGAAUCGAAAAGCAAAUGCUACCCAUUAACAAUAAAAUACACA-GCGAAAAAACAGUGGGGGUGGCAAAUAGGCUGGUCGAAGAGUGGGCGAAAUGUUUG---------- ....-.........((((..(((...((((((((.(((....)))....(((.-...........))).)))))))).....)))..))))...................---------- ( -22.80) >DroEre_CAF1 17000 107 - 1 GGAA-AAAAUUGGAUCGAAAAGCAAAUGCUACCCAUUAACAAUAAAAUACACA-GCGAAAAAACAGUGGGGGUGGCAAAUAGGCUGGUCGAAGAGUGGGCGAAAUGGGU----------- ....-.........((((..(((...((((((((.(((....)))....(((.-...........))).)))))))).....)))..))))..................----------- ( -22.80) >DroYak_CAF1 56101 107 - 1 GGAA-AAAAUUGGAUCGAAAAGCAAAUGCUACCCAUUAACAAUAAAAUACACA-GCGAAAAAACAGUGGGGGUGGCA-GUAGGCUGGUCGAAGAGUAGGCGAAAUGGUUU---------- ....-.........((((..(((...((((((((.(((....)))....(((.-...........))).))))))))-....)))..))))...................---------- ( -24.50) >consensus GAAA_AAAAUUGAAUCGAAAAGCAAAUGCUACCCAUUAACAAUAAAAUACACA_GCGAAAAAACAGUGGGGGUGGCAAAUAGGCUGGUCGAAGAGUGGGCGAAAUGGUUU__________ ..............((((..(((...((((((((...................................)))))))).....)))..))))............................. (-16.23 = -16.95 + 0.72)


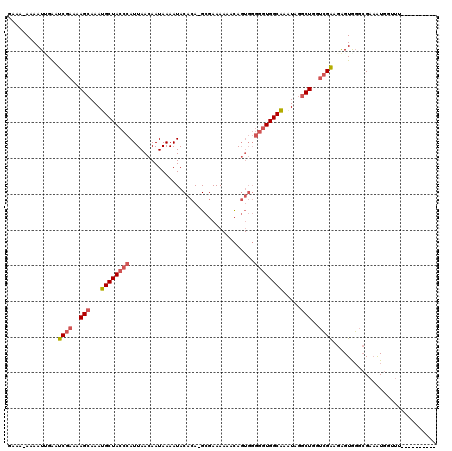
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:27 2006