
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,806,559 – 15,806,672 |
| Length | 113 |
| Max. P | 0.984069 |

| Location | 15,806,559 – 15,806,672 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -18.85 |
| Consensus MFE | -10.25 |
| Energy contribution | -10.70 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
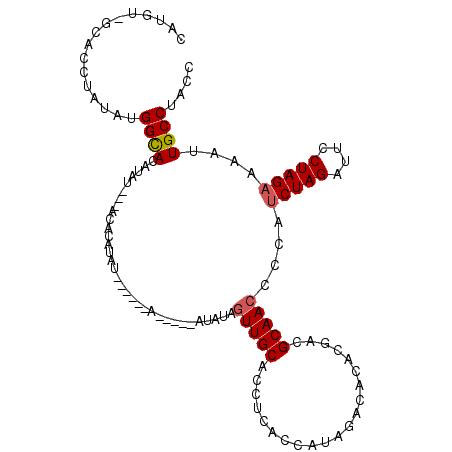
>3L_DroMel_CAF1 15806559 113 + 23771897 CAUGU-GCACCUAUAUGGUACAUAUAGUACACAUACAUAUGUAUAUGUAUAGAGUUGCACCUCACCAUAGACACACGACGCAAACCCAUCUAGAUUCCUAGAAAAUUGCCUACC ...((-(((((((((((((((((((.(((....))))))))))).))))))).).)))))...................((((.....(((((....)))))...))))..... ( -25.50) >DroAna_CAF1 3388 98 + 1 UAUAUUCCACACAUAUGGCAUAUAA---A-AUAUAU------AC----UCGUUGUUGCACCUCACCGCAGUCACA--ACGCAACCCCAUCUAGAUUCCUAGAAAAUUGCCUACC ................((((.....---.-......------..----.(((((((((........)))...)))--)))........(((((....)))))....)))).... ( -17.60) >DroYak_CAF1 3519 98 + 1 CAUGU-GCACCUAUAUGGCACAUAU---ACACAUAU------------AUCUAGUUGCACCUCACCAUAGACACACGACGCAACCCCAACUAGAUUCCUAGAAAAUUGCCUACC .((((-((.........))))))..---........------------(((((((((.............................)))))))))................... ( -13.45) >consensus CAUGU_GCACCUAUAUGGCACAUAU___ACACAUAU______A_____AUAUAGUUGCACCUCACCAUAGACACACGACGCAACCCCAUCUAGAUUCCUAGAAAAUUGCCUACC ................((((.................................(((((.....................)))))....(((((....)))))....)))).... (-10.25 = -10.70 + 0.45)

| Location | 15,806,559 – 15,806,672 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -16.45 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15806559 113 - 23771897 GGUAGGCAAUUUUCUAGGAAUCUAGAUGGGUUUGCGUCGUGUGUCUAUGGUGAGGUGCAACUCUAUACAUAUACAUAUGUAUGUGUACUAUAUGUACCAUAUAGGUGC-ACAUG .((((((.....(((((....)))))...))))))...(((..((((((....((((((......(((((((((....))))))))).....)))))).))))))..)-))... ( -39.40) >DroAna_CAF1 3388 98 - 1 GGUAGGCAAUUUUCUAGGAAUCUAGAUGGGGUUGCGU--UGUGACUGCGGUGAGGUGCAACAACGA----GU------AUAUAU-U---UUAUAUGCCAUAUGUGUGGAAUAUA .....((((((((((((....)))))..)))))))((--(((...((((......)))))))))..----..------.(((((-(---((((((.......)))))))))))) ( -24.10) >DroYak_CAF1 3519 98 - 1 GGUAGGCAAUUUUCUAGGAAUCUAGUUGGGGUUGCGUCGUGUGUCUAUGGUGAGGUGCAACUAGAU------------AUAUGUGU---AUAUGUGCCAUAUAGGUGC-ACAUG .....(((((((.((((....))))...)))))))...(((..((((((....((..((..((.((------------....)).)---)..))..)).))))))..)-))... ( -28.00) >consensus GGUAGGCAAUUUUCUAGGAAUCUAGAUGGGGUUGCGUCGUGUGUCUAUGGUGAGGUGCAACUACAU_____U______AUAUGUGU___AUAUGUGCCAUAUAGGUGC_ACAUG ((((.((((((((((((....)))))..)))))))....(((..((......))..)))...................................))))................ (-16.45 = -16.90 + 0.45)


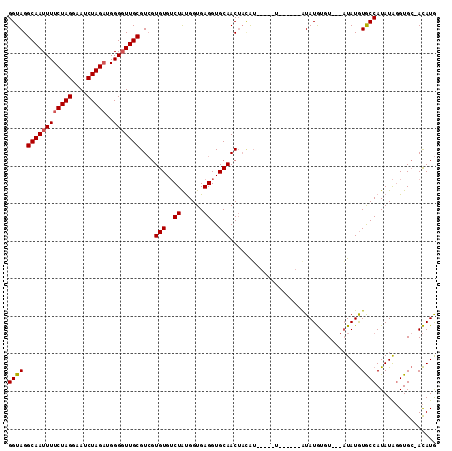
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:19 2006