
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,796,762 – 15,796,862 |
| Length | 100 |
| Max. P | 0.809459 |

| Location | 15,796,762 – 15,796,862 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -20.03 |
| Consensus MFE | -14.68 |
| Energy contribution | -14.80 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
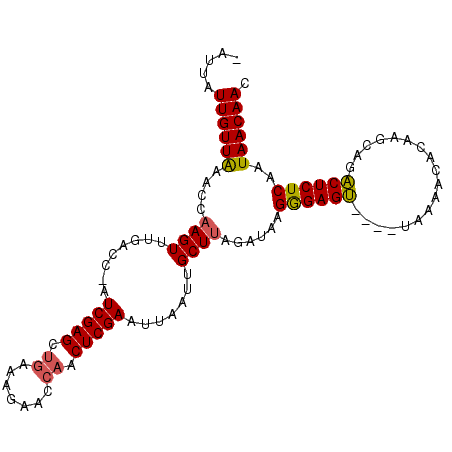
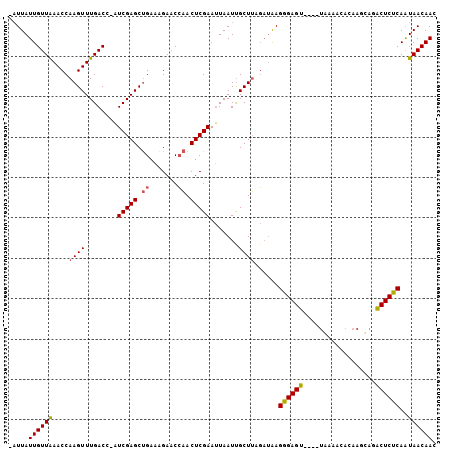
>3L_DroMel_CAF1 15796762 100 + 23771897 -CUUAUUGUUAAACCAAGUUUGACC-AUCGAGCUGAAUGAACCAACUCGAAUUAAUCGCUUAGAUAAGGGAGU----UAAAACACAAGUAGACUCUCAAUAACAAC -....((((((....((((......-.(((((.((.......)).))))).......))))......((((((----(...((....)).)))))))..)))))). ( -18.84) >DroAna_CAF1 816 106 + 1 UAUUAUUGUUGAACCAAGUUUGACCCUUCGAGCCGAUGGAACGUACUCGAAAUAAUUGCUAACCGGAGGGAGAACCCAAAAAAAAAAGGAGUCUCUCAGUAACAAC .....((((((..((.(((.......((((((.((......))..))))))......)))....)).((((((..((..........))..))))))..)))))). ( -20.72) >DroYak_CAF1 1037 100 + 1 -AUUAUUGUUAAACCAAGUUUGACC-AUCGAGCUGAAAGAACCAACUCGAAUUAAUUGCUUAGAUAAGAGAGU----UGAAACACAACCAGACUCUCAAUAACAAC -....((((((....((((......-.(((((.((.......)).))))).......))))......((((((----(............)))))))..)))))). ( -20.54) >consensus _AUUAUUGUUAAACCAAGUUUGACC_AUCGAGCUGAAAGAACCAACUCGAAUUAAUUGCUUAGAUAAGGGAGU____UAAAACACAAGCAGACUCUCAAUAACAAC .....((((((....((((........(((((.((.......)).))))).......))))......((((((..................))))))..)))))). (-14.68 = -14.80 + 0.12)

| Location | 15,796,762 – 15,796,862 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -15.54 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
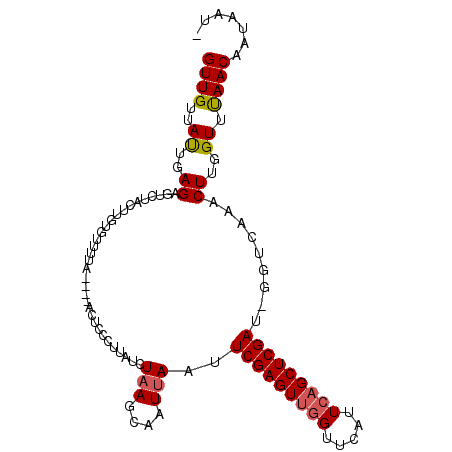
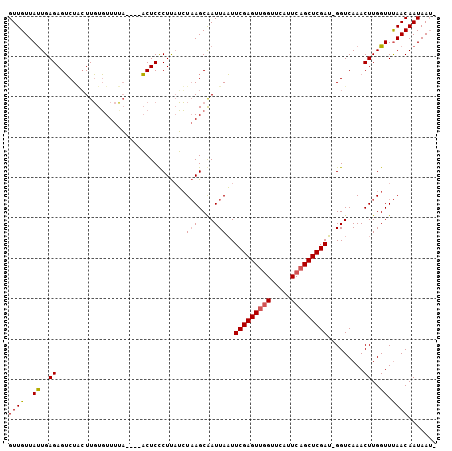
>3L_DroMel_CAF1 15796762 100 - 23771897 GUUGUUAUUGAGAGUCUACUUGUGUUUUA----ACUCCCUUAUCUAAGCGAUUAAUUCGAGUUGGUUCAUUCAGCUCGAU-GGUCAAACUUGGUUUAACAAUAAG- (((((((....((((..((....))....----))))......(((((.(((((..(((((((((.....))))))))))-))))...)))))..)))))))...- ( -24.40) >DroAna_CAF1 816 106 - 1 GUUGUUACUGAGAGACUCCUUUUUUUUUUGGGUUCUCCCUCCGGUUAGCAAUUAUUUCGAGUACGUUCCAUCGGCUCGAAGGGUCAAACUUGGUUCAACAAUAAUA ((((..((..((.(((((...........(((....)))................(((((((.((......))))))))))))))...))..)).))))....... ( -27.70) >DroYak_CAF1 1037 100 - 1 GUUGUUAUUGAGAGUCUGGUUGUGUUUCA----ACUCUCUUAUCUAAGCAAUUAAUUCGAGUUGGUUCUUUCAGCUCGAU-GGUCAAACUUGGUUUAACAAUAAU- (((((((.((((((...(((((.....))----))))))))).(((((........(((((((((.....))))))))).-.......)))))..)))))))...- ( -28.69) >consensus GUUGUUAUUGAGAGUCUACUUGUGUUUUA____ACUCCCUUAUCUAAGCAAUUAAUUCGAGUUGGUUCAUUCAGCUCGAU_GGUCAAACUUGGUUUAACAAUAAU_ ((((..((..((................................(((....)))..(((((((((.....))))))))).........))..)).))))....... (-15.54 = -16.10 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:15 2006