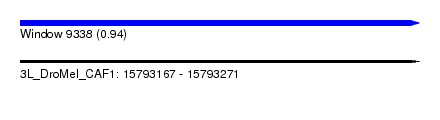
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,793,167 – 15,793,271 |
| Length | 104 |
| Max. P | 0.936643 |

| Location | 15,793,167 – 15,793,271 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.85 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -28.99 |
| Energy contribution | -29.10 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15793167 104 + 23771897 UCACUAUUCUACUA----UAUACUACCAUUUCUGCC--CACAGAUAGACGCCGUGUGGGGUUGGGUUGCUUCCGGCCCGCUGCGCUUAGCAGCGAACAGAAUGGGCGACC------ ...(((((((....----(((((.......((((..--..))))........)))))((((((((.....))))))))(((((.....)))))....)))))))......------ ( -34.36) >DroVir_CAF1 19655 92 + 1 --------------------GU-GUACUUUUUCAUCAAUAUAGAUAGACGCCGUGUGGGGUUGGG---CUUCCGCCCCGCUGCGCUUAGCGGCGAACAGAAUGGGCGAUCCUUUAC --------------------..-...((..((((((......)))...(((((((((.(((.(((---(....)))).))).)))...))))))....)))..))........... ( -29.10) >DroGri_CAF1 16663 98 + 1 ------UUCGGAUU---------UUUUGUUUUUGCCAAUUUAGAUAGACGCCGUGUGGGGUUGGG---CUUCCGCCCCGCUGCGCUUAGCGGCGAACAGAAUGGGCGAUCCUUUAC ------...(((((---------..........(((.((((.(.....(((((((((.(((.(((---(....)))).))).)))...))))))..).)))).))))))))..... ( -32.80) >DroSim_CAF1 47961 108 + 1 UCACUAUUCUACUUUAUAUGUACUUACAUUUCUGCC--CACAGAUAGACGCCGUGUGGGGUUGGGUUGCUUCCGGCCCGCUGCGCUUAGCAGCGAACAGAAUGGGCGACC------ .......((((......((((....)))).((((..--..))))))))((((.(.((((((((((.....))))))))(((((.....)))))...)).)...))))...------ ( -34.90) >DroEre_CAF1 43095 93 + 1 GCACUA---------------ACUACCAUUUCUACC--CACAGAUAGACGCCGUGUGGGGUUGGGUUGCUUCCGGCCCGCUGCGCUUAGCAGCGAACAGAAUGGGCGACC------ ......---------------....((((((...((--(((((........).))))))(((((((((....))))))(((((.....))))).))).))))))......------ ( -33.00) >DroAna_CAF1 36964 93 + 1 --------CAACUUUGUAUGUUUUUCCUUU---------CCAGAUAGACGCCGUGUGGGGUUGGGUUGCUUCCGGCCCGCUGCGUUUAGCAGCGAACAGAAUGGGCGAUC------ --------......................---------.........((((.(.((((((((((.....))))))))(((((.....)))))...)).)...))))...------ ( -30.70) >consensus ________C_ACUU______UACUUCCAUUUCUGCC__CACAGAUAGACGCCGUGUGGGGUUGGGUUGCUUCCGGCCCGCUGCGCUUAGCAGCGAACAGAAUGGGCGACC______ ................................................((((.(.((((((((((.....))))))))(((((.....)))))...)).)...))))......... (-28.99 = -29.10 + 0.11)

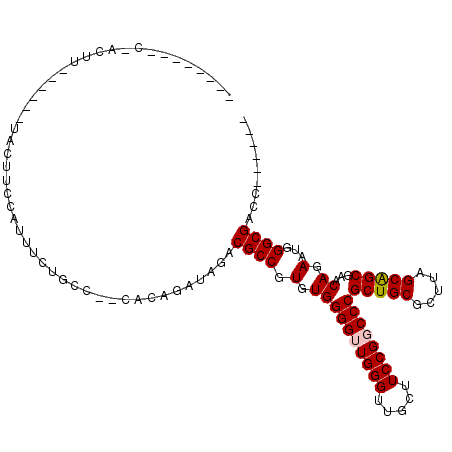
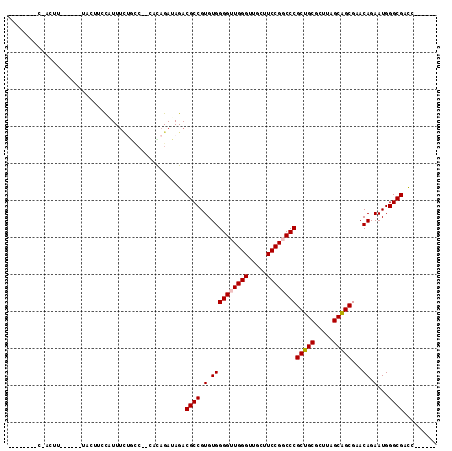
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:12 2006