| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,770,315 – 15,770,424 |
| Length | 109 |
| Max. P | 0.996539 |

| Location | 15,770,315 – 15,770,424 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -24.04 |
| Energy contribution | -26.88 |
| Covariance contribution | 2.84 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
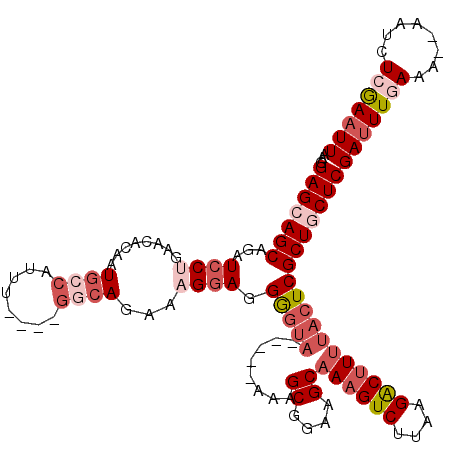
>3L_DroMel_CAF1 15770315 109 + 23771897 AAGGAGCAGCAGAUCCUAAACACAAUGGCAUUU----GGCAGAAAGGAGGGGUA-----AAAGCGGAAGCAAAGUCUUAAGACUUUUACUCGCUGCUCGAUUUGAAA--AAUCUCGAAUU ...(((((((...((((.....(((......))----)......)))).(((((-----(((((....))...(((....))))))))))))))))))(((((((..--....))))))) ( -36.60) >DroSec_CAF1 21919 109 + 1 AAGGAGCAGCAGAUCCCGAACACAAUGCCAUUU----GGCAGAAAGGAUGGGUA-----AAAGCGGAAGCAAAGUCUUAAGACUUUUACUCGCUGCUCGAUUUGAAA--AAUCUCGAAUU ...(((((((..((((.........((((....----))))....))))(((((-----(((((....))...(((....))))))))))))))))))(((((((..--....))))))) ( -39.72) >DroSim_CAF1 22989 109 + 1 AAGGAGCAGCAGAUCCUGAACACAAUGCCAUUU----GGCAGAAAGGAGGGGUA-----AAAGCGGAAGCAAAGUCUUAAGACUUUUACUCGCUGCUCGAUUUGAAA--AAUCUCGAAUU ...(((((((...((((........((((....----))))...)))).(((((-----(((((....))...(((....))))))))))))))))))(((((((..--....))))))) ( -41.30) >DroEre_CAF1 22210 109 + 1 AAGGAGCAGCAGAUCCUGAAUACAAUGCCAUUU----GGCAGAAAGGAGGGUUA-----GAAGCGGAAGCAAAGUCUUAAGGCUUUUACUCGCUACUCGAUUUAAAA--AAUCUCAAAUU ...(((.(((...((((........((((....----))))...))))((((..-----...((....))((((((....)))))).))))))).)))((((.....--))))....... ( -29.40) >DroAna_CAF1 18188 117 + 1 AAGGAGCAGCAGAUACGGACAACAAUA--AACGGGCCGUAAGAAAGGAUGAGGAGCUCGAAAGCGGAAGCAAAGUCUUAAGACCU-AGCUCGCUUCUCGAUUUAAAAAAAAACUAAAUUU ...(((.(((...(((((....(....--...)..)))))............(((((.(...((....))...(((....)))).-)))))))).)))...................... ( -26.60) >consensus AAGGAGCAGCAGAUCCUGAACACAAUGCCAUUU____GGCAGAAAGGAGGGGUA_____AAAGCGGAAGCAAAGUCUUAAGACUUUUACUCGCUGCUCGAUUUGAAA__AAUCUCGAAUU ...(((((((...((((........((((........))))...)))).(((((........((....))((((((....))))))))))))))))))(((((((........))))))) (-24.04 = -26.88 + 2.84)


| Location | 15,770,315 – 15,770,424 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.30 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -17.96 |
| Energy contribution | -18.92 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
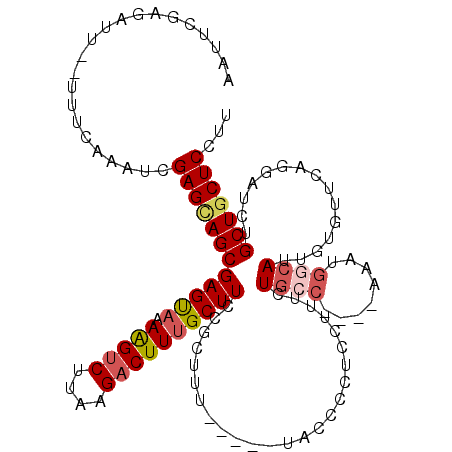
>3L_DroMel_CAF1 15770315 109 - 23771897 AAUUCGAGAUU--UUUCAAAUCGAGCAGCGAGUAAAAGUCUUAAGACUUUGCUUCCGCUUU-----UACCCCUCCUUUCUGCC----AAAUGCCAUUGUGUUUAGGAUCUGCUGCUCCUU .......((((--.....))))((((((((((((((.(((....)))))))))).......-----......((((....(((----((......))).))..))))...)))))))... ( -27.20) >DroSec_CAF1 21919 109 - 1 AAUUCGAGAUU--UUUCAAAUCGAGCAGCGAGUAAAAGUCUUAAGACUUUGCUUCCGCUUU-----UACCCAUCCUUUCUGCC----AAAUGGCAUUGUGUUCGGGAUCUGCUGCUCCUU .......((((--.....))))((((((((((((((.(((....)))))))))).......-----..(((........((((----....))))........)))....)))))))... ( -31.79) >DroSim_CAF1 22989 109 - 1 AAUUCGAGAUU--UUUCAAAUCGAGCAGCGAGUAAAAGUCUUAAGACUUUGCUUCCGCUUU-----UACCCCUCCUUUCUGCC----AAAUGGCAUUGUGUUCAGGAUCUGCUGCUCCUU .......((((--.....))))((((((((((((((.(((....)))))))))).......-----......((((..(((((----....))))....)...))))...)))))))... ( -28.90) >DroEre_CAF1 22210 109 - 1 AAUUUGAGAUU--UUUUAAAUCGAGUAGCGAGUAAAAGCCUUAAGACUUUGCUUCCGCUUC-----UAACCCUCCUUUCUGCC----AAAUGGCAUUGUAUUCAGGAUCUGCUGCUCCUU .......((((--.....))))(((((((((((..((((...........))))..)))).-----....(((......((((----....))))........)))....)))))))... ( -23.34) >DroAna_CAF1 18188 117 - 1 AAAUUUAGUUUUUUUUUAAAUCGAGAAGCGAGCU-AGGUCUUAAGACUUUGCUUCCGCUUUCGAGCUCCUCAUCCUUUCUUACGGCCCGUU--UAUUGUUGUCCGUAUCUGCUGCUCCUU .....((((...........((((((.((((((.-(((((....))))).)))..)))))))))................(((((..((..--......)).)))))...))))...... ( -23.50) >consensus AAUUCGAGAUU__UUUCAAAUCGAGCAGCGAGUAAAAGUCUUAAGACUUUGCUUCCGCUUU_____UACCCCUCCUUUCUGCC____AAAUGGCAUUGUGUUCAGGAUCUGCUGCUCCUU ......................((((((((((((.(((((....)))))))))).........................((((........))))...............)))))))... (-17.96 = -18.92 + 0.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:07 2006