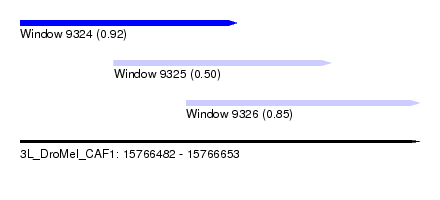
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,766,482 – 15,766,653 |
| Length | 171 |
| Max. P | 0.924188 |

| Location | 15,766,482 – 15,766,575 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -23.90 |
| Energy contribution | -24.15 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15766482 93 + 23771897 AGCAGCAACUAAACAUCAAUAAGCAUUGACUAACUGGAUUCGCAUUCCGGGCACAAGGACAACAACAACAA-------------------------CUCCCCAACUGGGGAGCACUCC-- ....((.........(((((....)))))....(((((.......)))))))...................-------------------------((((((....))))))......-- ( -21.60) >DroSec_CAF1 18075 116 + 1 AGCAGCAACUAAACAUCAAUAAGCAUUGACUAACUGGAUUCGCAUUCCGGGCACAAGGACAACAACAACAACAACAGC---AAGCGAAGGAUACGGCUCCCCAACUGGGGAGCACUCC-C .((.((.........(((((....)))))....(((((.......)))))))....(.....).............))---.......(((....(((((((....)))))))..)))-. ( -29.50) >DroSim_CAF1 19715 120 + 1 AGCAGCAACUAAACAUCAAUAAGCAUUGACUAACUGGAUUCGCAUUCCGGGCACAAGGACAACAACAACAACAACAGCAGUAAGCGAAGGAUACGGCUCCCCAACUGGGGAGCACUCCCC ....((.........(((((....)))))....(((((.......)))))))....((..................((.....))...(((....(((((((....)))))))..))))) ( -29.70) >DroEre_CAF1 18466 106 + 1 AGCAGCAACUAAACAUCAAUAAGCAUUGACUAACUGGAUUCGCAUUCCGGGCACAAGGACAACAACAAG------------AGGCGACGGAUACGGCUCCCCCACUGGGGAGCACUCC-- ....((.........(((((....)))))....(((((.......)))))))................(------------((....((....))(((((((....))))))).))).-- ( -28.50) >consensus AGCAGCAACUAAACAUCAAUAAGCAUUGACUAACUGGAUUCGCAUUCCGGGCACAAGGACAACAACAACAA__________AAGCGAAGGAUACGGCUCCCCAACUGGGGAGCACUCC__ ....((.........(((((....)))))....(((((.......)))))))...........................................(((((((....)))))))....... (-23.90 = -24.15 + 0.25)


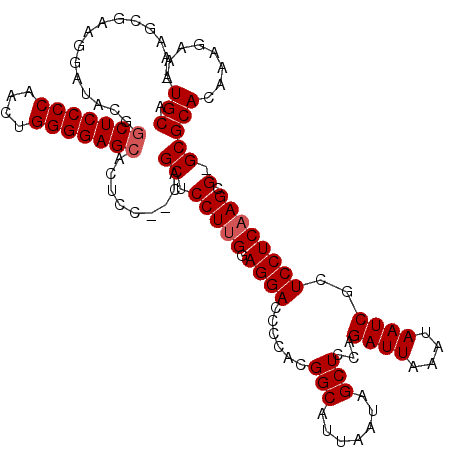
| Location | 15,766,522 – 15,766,615 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.43 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -26.21 |
| Energy contribution | -26.78 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15766522 93 + 23771897 CGCAUUCCGGGCACAAGGACAACAACAACAA-------------------------CUCCCCAACUGGGGAGCACUCC--CUGCUCCUUGGAGGACCCCACGGCAUUAAUAGCUUCAGAU .((.....((((.((((((............-------------------------.(((((....)))))(((....--.)))))))))...).)))....))................ ( -26.00) >DroSec_CAF1 18115 116 + 1 CGCAUUCCGGGCACAAGGACAACAACAACAACAACAGC---AAGCGAAGGAUACGGCUCCCCAACUGGGGAGCACUCC-CCAGCUCCUUGGAGGACCCCACGGCAUUAAUAGCUCCAGAU .((.....((((.((((((...................---..((...(((....(((((((....)))))))..)))-...))))))))...).)))....))................ ( -33.49) >DroSim_CAF1 19755 120 + 1 CGCAUUCCGGGCACAAGGACAACAACAACAACAACAGCAGUAAGCGAAGGAUACGGCUCCCCAACUGGGGAGCACUCCCCCAGCUCCUUGGAGGACCCCACGGCAUUAAUAGCUCCAGAU .((.....((((.(((((((................((.....))...(((....(((((((....)))))))..)))....).))))))...).)))....))................ ( -35.10) >DroEre_CAF1 18506 106 + 1 CGCAUUCCGGGCACAAGGACAACAACAAG------------AGGCGACGGAUACGGCUCCCCCACUGGGGAGCACUCC--CCGCUCCUCGGAGGACCCCACGGCAUUAAUAGCUCCAGAU .....((.((((................(------------(((.(.(((.....(((((((....))))))).....--)))).))))((......))............)).)).)). ( -33.80) >consensus CGCAUUCCGGGCACAAGGACAACAACAACAA__________AAGCGAAGGAUACGGCUCCCCAACUGGGGAGCACUCC__CAGCUCCUUGGAGGACCCCACGGCAUUAAUAGCUCCAGAU .((.....((((.(((((((........................(....).....(((((((....))))))).........).))))))...).)))....))................ (-26.21 = -26.78 + 0.56)


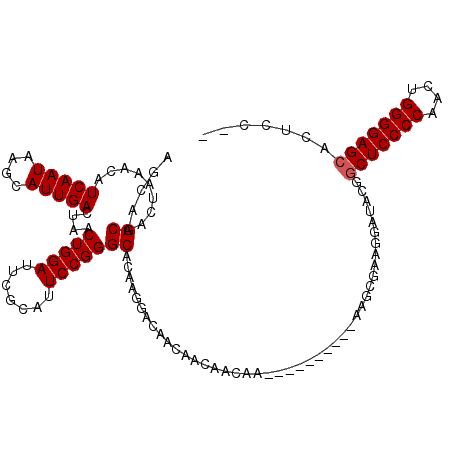
| Location | 15,766,553 – 15,766,653 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.20 |
| Mean single sequence MFE | -34.97 |
| Consensus MFE | -27.85 |
| Energy contribution | -28.35 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15766553 100 + 23771897 ----------------CUCCCCAACUGGGGAGCACUCC--CUGCUCCUUGGAGGACCCCACGGCAUUAAUAGCUUCAGAUUAAAUAAUCGCUCCUCAAGCG-GCGCACAAAGAAAUGCA ----------------((((((....))))))......--..(((.((((.((((......(((.......)))...((((....))))..)))))))).)-))(((........))). ( -28.30) >DroSec_CAF1 18153 116 + 1 -AAGCGAAGGAUACGGCUCCCCAACUGGGGAGCACUCC-CCAGCUCCUUGGAGGACCCCACGGCAUUAAUAGCUCCAGAUUAAAUAAUCGCUCCUCAAGCG-GCGCACAAAGAAAUGCA -..((...(((....(((((((....)))))))..)))-...(((.((((.((((......(((.......)))...((((....))))..)))))))).)-))))............. ( -36.30) >DroSim_CAF1 19795 118 + 1 UAAGCGAAGGAUACGGCUCCCCAACUGGGGAGCACUCCCCCAGCUCCUUGGAGGACCCCACGGCAUUAAUAGCUCCAGAUUAAAUAAUCGCUCCUCAAGCG-GCGCACAAAGAAAUGCA ...((...(((....(((((((....)))))))..)))....(((.((((.((((......(((.......)))...((((....))))..)))))))).)-))))............. ( -36.30) >DroEre_CAF1 18535 116 + 1 -AGGCGACGGAUACGGCUCCCCCACUGGGGAGCACUCC--CCGCUCCUCGGAGGACCCCACGGCAUUAAUAGCUCCAGAUUAAAUAAUCGCUCCUCAAGCGGGCGCACAAAGAAAUGCA -..(((.((....))(((((((....)))))))....(--(((((.....(((((......(((.......)))...((((....))))..))))).)))))))))............. ( -39.00) >consensus _AAGCGAAGGAUACGGCUCCCCAACUGGGGAGCACUCC__CAGCUCCUUGGAGGACCCCACGGCAUUAAUAGCUCCAGAUUAAAUAAUCGCUCCUCAAGCG_GCGCACAAAGAAAUGCA ...............(((((((....))))))).........((.(((((.((((......(((.......)))...((((....))))..)))))))).).))(((........))). (-27.85 = -28.35 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:02:00 2006