
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,764,557 – 15,764,793 |
| Length | 236 |
| Max. P | 0.953139 |

| Location | 15,764,557 – 15,764,673 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -29.25 |
| Energy contribution | -29.62 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
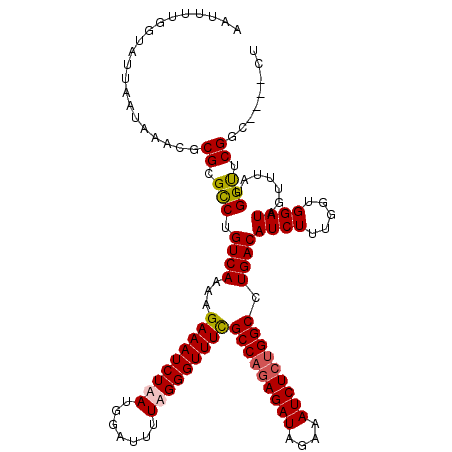
>3L_DroMel_CAF1 15764557 116 + 23771897 AAUUUUGGUAUUAAUAAACGCGCGUCUGUCAAAAGAAAUCUAAUGGAUUUUAGGGUUUUGCCAGAGAUAGAAAUCUCUGGCCUGACAUCUUUGGUGGAUAGUUUAUGGCUCGGC----CU ......(((....((((((....(((..(((((.((((((.....))))))...(((..(((((((((....)))))))))..)))...)))))..))).))))))......))----). ( -37.00) >DroSec_CAF1 16136 116 + 1 AAUUUUGGUAUUAAUAAACGCGCGCCUGUCAAAAGAAAUCUAAUGGAUUUUUGGGUUUCGCCAGAGAUAGAAAUCUCUGGCCUGACAUCUUUGGUGGAUAGUUUAUGGUUCGGC----CU ......(((....((((((.(.(((((((((...(((((((((.......)))))))))(((((((((....))))))))).))))).....)))))...))))))......))----). ( -40.10) >DroSim_CAF1 17136 116 + 1 AAUUUUGGUAUUAAUAAACGCGCGCCUGUCAAAAGAAAUCUAAUGGAUUUUUGGGUUUCGCCAGAGAUAGAAAUCUCUGGCCUGACAUCUUUGGUGGAUAGUUUAUGGUUCGGC----CU ......(((....((((((.(.(((((((((...(((((((((.......)))))))))(((((((((....))))))))).))))).....)))))...))))))......))----). ( -40.10) >DroEre_CAF1 16254 120 + 1 AAUUUUGGUAUUAAUAAACGCGCGCCUGUCAAAAGAAAUCUAAUGGAUUUUAGGGUUUUGCCUGAGAUAGAAAUCUGUGGCCUGACAUCUUUGCUGGAUAGUUUAUGGUUCGGCUUGGCU .......................((((((((..((((..((((......))))..))))(((..((((....))))..))).))))).....(((((((........)))))))..))). ( -26.80) >consensus AAUUUUGGUAUUAAUAAACGCGCGCCUGUCAAAAGAAAUCUAAUGGAUUUUAGGGUUUCGCCAGAGAUAGAAAUCUCUGGCCUGACAUCUUUGGUGGAUAGUUUAUGGUUCGGC____CU ....................((.(((.((((...(((((((((.......)))))))))(((((((((....))))))))).))))((((.....)))).......))).))........ (-29.25 = -29.62 + 0.38)

| Location | 15,764,557 – 15,764,673 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -21.98 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15764557 116 - 23771897 AG----GCCGAGCCAUAAACUAUCCACCAAAGAUGUCAGGCCAGAGAUUUCUAUCUCUGGCAAAACCCUAAAAUCCAUUAGAUUUCUUUUGACAGACGCGCGUUUAUUAAUACCAAAAUU .(----((...)))((((((.(((.......)))((((((((((((((....)))))))))......((((......)))).......)))))........))))))............. ( -26.40) >DroSec_CAF1 16136 116 - 1 AG----GCCGAACCAUAAACUAUCCACCAAAGAUGUCAGGCCAGAGAUUUCUAUCUCUGGCGAAACCCAAAAAUCCAUUAGAUUUCUUUUGACAGGCGCGCGUUUAUUAAUACCAAAAUU ((----((((..((.......(((.......)))((((((((((((((....))))))))).........(((((.....)))))...))))).))..)).))))............... ( -26.70) >DroSim_CAF1 17136 116 - 1 AG----GCCGAACCAUAAACUAUCCACCAAAGAUGUCAGGCCAGAGAUUUCUAUCUCUGGCGAAACCCAAAAAUCCAUUAGAUUUCUUUUGACAGGCGCGCGUUUAUUAAUACCAAAAUU ((----((((..((.......(((.......)))((((((((((((((....))))))))).........(((((.....)))))...))))).))..)).))))............... ( -26.70) >DroEre_CAF1 16254 120 - 1 AGCCAAGCCGAACCAUAAACUAUCCAGCAAAGAUGUCAGGCCACAGAUUUCUAUCUCAGGCAAAACCCUAAAAUCCAUUAGAUUUCUUUUGACAGGCGCGCGUUUAUUAAUACCAAAAUU .((...(((............(((.......)))((((((((..((((....))))..)))......((((......)))).......))))).)))..))................... ( -19.20) >consensus AG____GCCGAACCAUAAACUAUCCACCAAAGAUGUCAGGCCAGAGAUUUCUAUCUCUGGCAAAACCCAAAAAUCCAUUAGAUUUCUUUUGACAGGCGCGCGUUUAUUAAUACCAAAAUU ......(((............(((.......)))((((((((((((((....))))))))).........(((((.....)))))...))))).)))....................... (-21.98 = -22.72 + 0.75)


| Location | 15,764,597 – 15,764,713 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.80 |
| Mean single sequence MFE | -34.67 |
| Consensus MFE | -29.11 |
| Energy contribution | -29.67 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15764597 116 + 23771897 UAAUGGAUUUUAGGGUUUUGCCAGAGAUAGAAAUCUCUGGCCUGACAUCUUUGGUGGAUAGUUUAUGGCUCGGC----CUUUGCAUCAGUGACUCACAUAUGAUUGUUGAGUCACAUUUG ......(((..((((((..(((((((((....)))))))))..))).)))..))).(((.((....(((...))----)...))))).((((((((((......)).))))))))..... ( -38.00) >DroSec_CAF1 16176 116 + 1 UAAUGGAUUUUUGGGUUUCGCCAGAGAUAGAAAUCUCUGGCCUGACAUCUUUGGUGGAUAGUUUAUGGUUCGGC----CUUUGCAUCAGUGACUCACAUAUGAUCGUUGAGUCACAUCUC .....(((.(..(((((..(((((((((....)))))))))..(((((((.....)))).)))........)))----))..).))).(((((((((........).))))))))..... ( -35.10) >DroSim_CAF1 17176 116 + 1 UAAUGGAUUUUUGGGUUUCGCCAGAGAUAGAAAUCUCUGGCCUGACAUCUUUGGUGGAUAGUUUAUGGUUCGGC----CUUUGCAUCAGUGACUCACAUAUGAUCGUUGAGGCACAUCUC ..(((((((.....(((..(((((((((....)))))))))..)))((((.....)))))))))))(((...((----(((.(((((((((.....))).)))).)).)))))..))).. ( -33.30) >DroEre_CAF1 16294 120 + 1 UAAUGGAUUUUAGGGUUUUGCCUGAGAUAGAAAUCUGUGGCCUGACAUCUUUGCUGGAUAGUUUAUGGUUCGGCUUGGCUUUGCCUCAGUGACUCACAUAUGAUCGUUGAGUCACAGCUG ......((((((((......)))))))).......((.(((.......((..(((((((........)))))))..))....))).))(((((((((........).))))))))..... ( -32.30) >consensus UAAUGGAUUUUAGGGUUUCGCCAGAGAUAGAAAUCUCUGGCCUGACAUCUUUGGUGGAUAGUUUAUGGUUCGGC____CUUUGCAUCAGUGACUCACAUAUGAUCGUUGAGUCACAUCUC ..(((((((.....(((..(((((((((....)))))))))..)))((((.....))))))))))).((..((.....))..))....(((((((((........).))))))))..... (-29.11 = -29.67 + 0.56)



| Location | 15,764,597 – 15,764,713 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.80 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -24.17 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15764597 116 - 23771897 CAAAUGUGACUCAACAAUCAUAUGUGAGUCACUGAUGCAAAG----GCCGAGCCAUAAACUAUCCACCAAAGAUGUCAGGCCAGAGAUUUCUAUCUCUGGCAAAACCCUAAAAUCCAUUA ..(((((((((((.((......)))))))))(((((.....(----((...))).......(((.......))))))))(((((((((....)))))))))..............)))). ( -32.70) >DroSec_CAF1 16176 116 - 1 GAGAUGUGACUCAACGAUCAUAUGUGAGUCACUGAUGCAAAG----GCCGAACCAUAAACUAUCCACCAAAGAUGUCAGGCCAGAGAUUUCUAUCUCUGGCGAAACCCAAAAAUCCAUUA ..(((((((((((.((......)))))))))(((((.....(----(.....)).......(((.......))))))))(((((((((....)))))))))..............)))). ( -29.70) >DroSim_CAF1 17176 116 - 1 GAGAUGUGCCUCAACGAUCAUAUGUGAGUCACUGAUGCAAAG----GCCGAACCAUAAACUAUCCACCAAAGAUGUCAGGCCAGAGAUUUCUAUCUCUGGCGAAACCCAAAAAUCCAUUA ..(((((((.(((..(((((....))).))..))).))...(----(.....))..................)))))..(((((((((....)))))))))................... ( -24.80) >DroEre_CAF1 16294 120 - 1 CAGCUGUGACUCAACGAUCAUAUGUGAGUCACUGAGGCAAAGCCAAGCCGAACCAUAAACUAUCCAGCAAAGAUGUCAGGCCACAGAUUUCUAUCUCAGGCAAAACCCUAAAAUCCAUUA ..(((((((((((.((......))))))))))...(((........)))................)))...........(((..((((....))))..)))................... ( -25.30) >consensus CAGAUGUGACUCAACGAUCAUAUGUGAGUCACUGAUGCAAAG____GCCGAACCAUAAACUAUCCACCAAAGAUGUCAGGCCAGAGAUUUCUAUCUCUGGCAAAACCCAAAAAUCCAUUA ..(((((((((((.((......)))))))))((((.((........))............((((.......))))))))(((((((((....)))))))))..............)))). (-24.17 = -24.80 + 0.63)


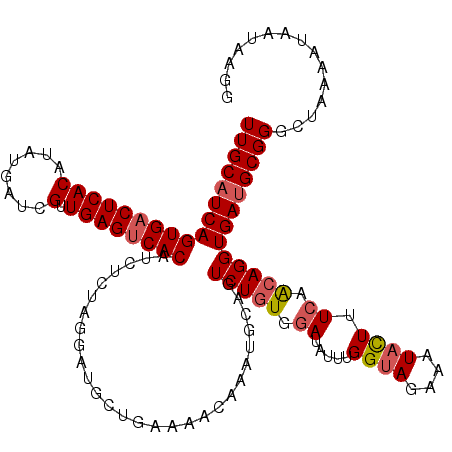
| Location | 15,764,673 – 15,764,793 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -22.59 |
| Energy contribution | -22.77 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.581050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15764673 120 + 23771897 UUGCAUCAGUGACUCACAUAUGAUUGUUGAGUCACAUUUGUAGGAUGCUGAAAACAAAUGCAGUCUAUGGAUAUUUGGUAGAAAUACUUUCAACAGGUGAUGCGGGCUAAAAUAAUAAGA ((((((((((((((((((......)).))))))))(((..(((..(((...........)))..)))..)))....((((....)))).........))))))))............... ( -35.10) >DroSec_CAF1 16252 120 + 1 UUGCAUCAGUGACUCACAUAUGAUCGUUGAGUCACAUCUCUAGGAUGCUGAAAACAAAUGCUGUCUGUGGAUAUUUGGUAGAAAUACUUUCAACAGGUGAUGCGGGCUAAAAUAAUAGGG (((((((((((((((((........).))))))))..........((.(((((.((((((((......)).))))))(((....)))))))).))..))))))))............... ( -30.70) >DroSim_CAF1 17252 120 + 1 UUGCAUCAGUGACUCACAUAUGAUCGUUGAGGCACAUCUCUAGGAUGCUGAAAACAAAUGGAGUCUGUGGAUAUUUGGUAGAAAUACUUUCAACAGGUGAUGCGGGCUAAAAUAAUAAGG ((((((((.((((((.(((.((....((..((((..((.....))))))..)).)).))))))))(((.((.....((((....)))).)).)))).))))))))............... ( -27.80) >DroEre_CAF1 16374 113 + 1 UUGCCUCAGUGACUCACAUAUGAUCGUUGAGUCACAGCUGUAGGAU-------ACAAAUGGAGUCUGUGAAUAUUUGGAAGGAAUAUUUUCAGCAGGUGAUGCGGGCUAAAAUUCUGCGG ........(((((((((........).))))))))..(((((((((-------........(((((((...((((((....(((....)))..))))))..)))))))...))))))))) ( -30.40) >consensus UUGCAUCAGUGACUCACAUAUGAUCGUUGAGUCACAUCUCUAGGAUGCUGAAAACAAAUGCAGUCUGUGGAUAUUUGGUAGAAAUACUUUCAACAGGUGAUGCGGGCUAAAAUAAUAAGG (((((((((((((((((........).))))))))............................(((((((((((((.....)))))..))).)))))))))))))............... (-22.59 = -22.77 + 0.19)


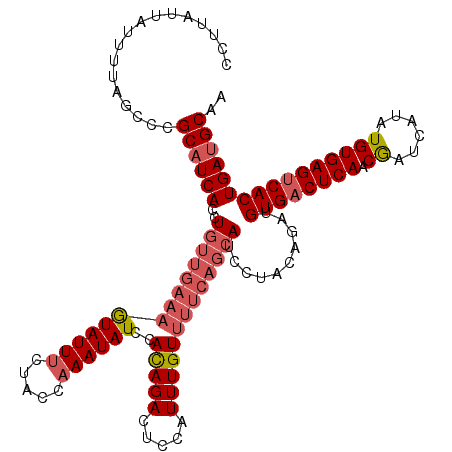
| Location | 15,764,673 – 15,764,793 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -20.16 |
| Energy contribution | -22.10 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15764673 120 - 23771897 UCUUAUUAUUUUAGCCCGCAUCACCUGUUGAAAGUAUUUCUACCAAAUAUCCAUAGACUGCAUUUGUUUUCAGCAUCCUACAAAUGUGACUCAACAAUCAUAUGUGAGUCACUGAUGCAA .................((((((..((((((((((((((.....))))))..(((((.....)))))))))))))..........((((((((.((......)))))))))))))))).. ( -27.00) >DroSec_CAF1 16252 120 - 1 CCCUAUUAUUUUAGCCCGCAUCACCUGUUGAAAGUAUUUCUACCAAAUAUCCACAGACAGCAUUUGUUUUCAGCAUCCUAGAGAUGUGACUCAACGAUCAUAUGUGAGUCACUGAUGCAA .................((((((..((((((((((((((.....))))))..(((((.....)))))))))))))..........((((((((.((......)))))))))))))))).. ( -27.70) >DroSim_CAF1 17252 120 - 1 CCUUAUUAUUUUAGCCCGCAUCACCUGUUGAAAGUAUUUCUACCAAAUAUCCACAGACUCCAUUUGUUUUCAGCAUCCUAGAGAUGUGCCUCAACGAUCAUAUGUGAGUCACUGAUGCAA .................((((((..(((.((...(((((.....))))))).)))(((((((((((((....(((((.....))))).....)))))....))).)))))..)))))).. ( -24.30) >DroEre_CAF1 16374 113 - 1 CCGCAGAAUUUUAGCCCGCAUCACCUGCUGAAAAUAUUCCUUCCAAAUAUUCACAGACUCCAUUUGU-------AUCCUACAGCUGUGACUCAACGAUCAUAUGUGAGUCACUGAGGCAA .............((((.........((((..((((((.......)))))).(((((.....)))))-------......)))).((((((((.((......)))))))))).).))).. ( -22.60) >consensus CCUUAUUAUUUUAGCCCGCAUCACCUGUUGAAAGUAUUUCUACCAAAUAUCCACAGACUCCAUUUGUUUUCAGCAUCCUACAGAUGUGACUCAACGAUCAUAUGUGAGUCACUGAUGCAA .................((((((..((((((((((((((.....))))))..(((((.....)))))))))))))..........((((((((.((......)))))))))))))))).. (-20.16 = -22.10 + 1.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:56 2006