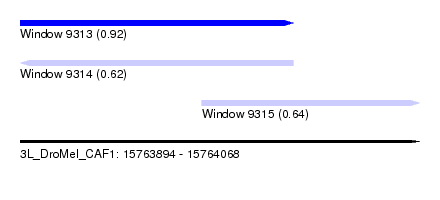
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,763,894 – 15,764,068 |
| Length | 174 |
| Max. P | 0.922456 |

| Location | 15,763,894 – 15,764,013 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.66 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -25.70 |
| Energy contribution | -28.68 |
| Covariance contribution | 2.98 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15763894 119 + 23771897 GGAUAUUCUU-UUCCCACAGUAGAAAGGAUAAUAAAAUGCCUACGGGCAAACAAAAAUAAACAAGAUACGUACGCCCGCUUGGAUGCAUCCUCAAAGGAUGGAUCCUUGAGGCAAGUCGA .(((..((((-(((........)))))))........(((((.(((((..((.................))..)))))(..((((.(((((.....))))).))))..)))))).))).. ( -35.93) >DroSec_CAF1 15466 120 + 1 GGAUAUUCUUUUUCCCACAGUAGAAAGGAUAAUAAAAUGCCUACGGGCAAACAAAAAUAAACAAGAUGCGUACGCCCGGUUGGAUGCAUCCUCAAAGGAUGGAUCCUUGAGGGAAGUCGA ...((((((((((........))))))))))......((((....))))...................((....(((....((((.(((((.....))))).))))....)))....)). ( -36.20) >DroSim_CAF1 16465 120 + 1 GGAUAUUCUUUUUCCCACAGUAGAAAGGAUAAUAAAAUGCCUACGGGCAAACAAAAAUAAACAAGAUGCGUACGCCCGGUUGGAUGCAUCCUCAAAGGAUGGAUCCUUGAGGGAAGUCGA ...((((((((((........))))))))))......((((....))))...................((....(((....((((.(((((.....))))).))))....)))....)). ( -36.20) >DroEre_CAF1 15603 119 + 1 GGAUAUUCUU-UUCCCACAGUAGAAAGGAUAAUAAAAUGCCUGCGGGCAAACAAAAAUAAACAAGAUGCGUACGACCGGUUGGGUGCAUCCUCAAAGGAUGGAUCCUUGAGGGAGGACGA ......((((-(((........)))))))........((((....))))...................(((.(..((....((((.(((((.....))))).))))....))..).))). ( -32.10) >DroAna_CAF1 10635 119 + 1 GGUUAUUCAUUUUCCCACACAAGAAAGGAUAAUAAAAUGCCUCGGGGCAAACAAAAAUAAACAACAUGCGUACGCCCGACUGGCUGCAUCCUUUAAGGAUGGAUCCUUGACGG-GGCCAA .(((((((.((((........)))).))))))).....((((((((((..((.................))..))))....((.(.(((((.....))))).).))....)))-)))... ( -30.43) >consensus GGAUAUUCUUUUUCCCACAGUAGAAAGGAUAAUAAAAUGCCUACGGGCAAACAAAAAUAAACAAGAUGCGUACGCCCGGUUGGAUGCAUCCUCAAAGGAUGGAUCCUUGAGGGAAGUCGA ...((((((((((........))))))))))......((((....))))...................((....(((....((((.(((((.....))))).))))....)))....)). (-25.70 = -28.68 + 2.98)


| Location | 15,763,894 – 15,764,013 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.66 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -23.60 |
| Energy contribution | -24.68 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15763894 119 - 23771897 UCGACUUGCCUCAAGGAUCCAUCCUUUGAGGAUGCAUCCAAGCGGGCGUACGUAUCUUGUUUAUUUUUGUUUGCCCGUAGGCAUUUUAUUAUCCUUUCUACUGUGGGAA-AAGAAUAUCC ..((..(((((...((((.(((((.....))))).))))..(((((((.(((...............))).))))))))))))..))....((.(((((......))))-).))...... ( -35.36) >DroSec_CAF1 15466 120 - 1 UCGACUUCCCUCAAGGAUCCAUCCUUUGAGGAUGCAUCCAACCGGGCGUACGCAUCUUGUUUAUUUUUGUUUGCCCGUAGGCAUUUUAUUAUCCUUUCUACUGUGGGAAAAAGAAUAUCC ((...(((((.((.((((.(((((.....))))).))))....(((((...(((.............))).)))))(((((...............))))))).)))))...))...... ( -32.18) >DroSim_CAF1 16465 120 - 1 UCGACUUCCCUCAAGGAUCCAUCCUUUGAGGAUGCAUCCAACCGGGCGUACGCAUCUUGUUUAUUUUUGUUUGCCCGUAGGCAUUUUAUUAUCCUUUCUACUGUGGGAAAAAGAAUAUCC ((...(((((.((.((((.(((((.....))))).))))....(((((...(((.............))).)))))(((((...............))))))).)))))...))...... ( -32.18) >DroEre_CAF1 15603 119 - 1 UCGUCCUCCCUCAAGGAUCCAUCCUUUGAGGAUGCACCCAACCGGUCGUACGCAUCUUGUUUAUUUUUGUUUGCCCGCAGGCAUUUUAUUAUCCUUUCUACUGUGGGAA-AAGAAUAUCC .........((((((((....))).)))))((((((((.....))).....))))).....(((((((.....(((((((....................)))))))..-)))))))... ( -28.35) >DroAna_CAF1 10635 119 - 1 UUGGCC-CCGUCAAGGAUCCAUCCUUAAAGGAUGCAGCCAGUCGGGCGUACGCAUGUUGUUUAUUUUUGUUUGCCCCGAGGCAUUUUAUUAUCCUUUCUUGUGUGGGAAAAUGAAUAACC ....((-(((.((((((..(((((.....)))))..(((..(((((.(((.(((.............))).))))))))))).............)))))))).)))............. ( -33.72) >consensus UCGACUUCCCUCAAGGAUCCAUCCUUUGAGGAUGCAUCCAACCGGGCGUACGCAUCUUGUUUAUUUUUGUUUGCCCGUAGGCAUUUUAUUAUCCUUUCUACUGUGGGAAAAAGAAUAUCC ((...(((((.((.((((.(((((.....))))).))))....(((((.(((...............))).)))))(((((...............))))))).)))))...))...... (-23.60 = -24.68 + 1.08)



| Location | 15,763,973 – 15,764,068 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -22.90 |
| Energy contribution | -22.78 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15763973 95 + 23771897 UGGAUGCAUCCUCAAAGGAUGGAUCCUUGAGGCAAGUCGAGGUGAAGGUUUAGAC-------------------------UUAGAUUGUGGCAUUGAUUUCAUUUUCAGCUGUCUAUCAA .((((.(((((.....))))).))))...(((((.((((..((..((((....))-------------------------))..))..)))).((((........)))).)))))..... ( -25.40) >DroSec_CAF1 15546 95 + 1 UGGAUGCAUCCUCAAAGGAUGGAUCCUUGAGGGAAGUCGAGGUGAAGGCUUGGAC-------------------------UUAGAUUGUGGCAUUGAUUUCAUUUUCAGCUGUCUAUCAA .((((.(((((.....))))).)))).......(((((.((((....)))).)))-------------------------)).(((...((((((((........)))).)))).))).. ( -30.80) >DroSim_CAF1 16545 95 + 1 UGGAUGCAUCCUCAAAGGAUGGAUCCUUGAGGGAAGUCGAGGUGAAGGCUUGGAC-------------------------UUAGAUUGUGGCAUUGAUUUCAUUUUCAGCUGUCUAUCAA .((((.(((((.....))))).)))).......(((((.((((....)))).)))-------------------------)).(((...((((((((........)))).)))).))).. ( -30.80) >DroEre_CAF1 15682 95 + 1 UGGGUGCAUCCUCAAAGGAUGGAUCCUUGAGGGAGGACGAGCUGCAGGUUUAGAC-------------------------UUAGAUUGUGGCAUUGAUUUCAUUUUCAGCUGUCUAUCAA .((((.(((((.....))))).)))).....((.(((((.((..(((.((((...-------------------------.)))))))..)).((((........)))).))))).)).. ( -27.10) >DroAna_CAF1 10715 119 + 1 UGGCUGCAUCCUUUAAGGAUGGAUCCUUGACGG-GGCCAAGGGAAAGGCUUAGAGACAAGCUCGGUGAUGCAGUGAUUGUUGGGAUUGUGGCAUUGAUUUCAUUUUCAGCUGUCUAUCAA ..(((((((((((((((((....)))))))..(-((((........))))).(((.....))))).))))))))(((.((((..(..((((........)))))..)))).)))...... ( -35.00) >consensus UGGAUGCAUCCUCAAAGGAUGGAUCCUUGAGGGAAGUCGAGGUGAAGGCUUAGAC_________________________UUAGAUUGUGGCAUUGAUUUCAUUUUCAGCUGUCUAUCAA .((((.(((((.....))))).)))).......(((((........)))))................................(((...((((((((........)))).)))).))).. (-22.90 = -22.78 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:49 2006