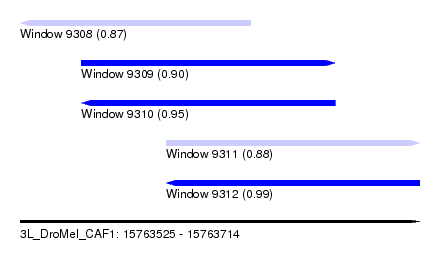
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,763,525 – 15,763,714 |
| Length | 189 |
| Max. P | 0.986262 |

| Location | 15,763,525 – 15,763,634 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -16.50 |
| Consensus MFE | -11.72 |
| Energy contribution | -12.24 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15763525 109 - 23771897 AUUUGAUUUUCCCUCAGCUGCCAAAGCCUUUUUGGCUUGUUUAUGCUAGAGAAAAUUGAAAUUUUAUGCAUUUCCCGCAUUUACCACCAAAA-----------ACCAAACCCCAAUUUCA .((..((((((.((.(((.....(((((.....)))))......))))).))))))..)).....((((.......))))............-----------................. ( -21.50) >DroSec_CAF1 15100 105 - 1 AUUUGAUUUUCCCUCGGCUGCCAAAGCCUUUUUGGCUUGUUUAUGCUACAGAAAAUUGAAAUUUUAUGCAUUUCCCC---------CCAAAAACCAA------ACCAAACCCCAAUUUCA .((..((((((....(((.....(((((.....)))))......)))...))))))..)).................---------...........------................. ( -16.80) >DroSim_CAF1 16095 105 - 1 AUUUCAUUUUCCCUCGGCUGCCAAAGCCUUUUUGGCUUGUUUAUGCUACAGAAAAUUGAAAUUUUAUGCAUUUCCCC---------CCAAAAACCAA------ACCAAACCCCAAUUUCA (((((((((((....(((.....(((((.....)))))......)))...))))).))))))...............---------...........------................. ( -15.60) >DroEre_CAF1 15245 97 - 1 AUUUGAUUUUCCCUCGGCUGCCAAAGCCUUUUUGGCUUGUUUAUGCUGGAGAAAAUUGAAAUUUUAUGCACAUCCCC-----------------CAA------ACCAAACCCCAAUUUCA .((..((((((.(.((((.....(((((.....)))))......))))).))))))..)).................-----------------...------................. ( -20.80) >DroAna_CAF1 10348 111 - 1 AUUUGAUUUUCCCUCGUCUGCCUAGAUCCCCAUACCUUGUUUAUCCUUGAGAAAAUUGAGAUUUUAUGCAUUUCCCU---------CCUCUACUCAAUCAUUUCCCAAAAACUAAGUUCA ....(((((...(......)...))))).......(((((((....(((.((((((((((.................---------......))))))..)))).))))))).))).... ( -7.80) >consensus AUUUGAUUUUCCCUCGGCUGCCAAAGCCUUUUUGGCUUGUUUAUGCUAGAGAAAAUUGAAAUUUUAUGCAUUUCCCC_________CCAAAA__CAA______ACCAAACCCCAAUUUCA .((..((((((....(((.....(((((.....)))))......)))...))))))..))............................................................ (-11.72 = -12.24 + 0.52)

| Location | 15,763,554 – 15,763,674 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.52 |
| Mean single sequence MFE | -27.49 |
| Consensus MFE | -20.76 |
| Energy contribution | -21.56 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15763554 120 + 23771897 AUGCGGGAAAUGCAUAAAAUUUCAAUUUUCUCUAGCAUAAACAAGCCAAAAAGGCUUUGGCAGCUGAGGGAAAAUCAAAUCAAGCCCAGAGGCAAAAUUUAAUUGCGUUUAACCUGAAAU ...(((((((((((..((((((..((((((((((((......(((((.....))))).....))).)))))))))........(((....))).))))))...)))))))..)))).... ( -32.50) >DroSec_CAF1 15128 117 + 1 ---GGGGAAAUGCAUAAAAUUUCAAUUUUCUGUAGCAUAAACAAGCCAAAAAGGCUUUGGCAGCCGAGGGAAAAUCAAAUCAAGCCCAGAGGCAAAAUUUAAUUGCGUUUAACCUGAAAU ---((..(((((((..((((((......................((((((.....)))))).(((..(((..............)))...))).))))))...)))))))..))...... ( -28.34) >DroSim_CAF1 16123 117 + 1 ---GGGGAAAUGCAUAAAAUUUCAAUUUUCUGUAGCAUAAACAAGCCAAAAAGGCUUUGGCAGCCGAGGGAAAAUGAAAUCAAGCCCAGAGGCAAAAUUUAAUUGCGUUUAACCUGAAAU ---((..(((((((....((((((.((((((...((......(((((.....))))).....))...)))))).))))))...(((....)))..........)))))))..))...... ( -31.10) >DroEre_CAF1 15265 117 + 1 ---GGGGAUGUGCAUAAAAUUUCAAUUUUCUCCAGCAUAAACAAGCCAAAAAGGCUUUGGCAGCCGAGGGAAAAUCAAAUCAAGCCCAGAGGCAAAAUUUAAUUGCGUUUAACCUGAAAU ---((..(..((((..((((((..(((((((((.((......(((((.....))))).....)).).))))))))........(((....))).))))))...))))..)..))...... ( -26.20) >DroAna_CAF1 10382 117 + 1 ---AGGGAAAUGCAUAAAAUCUCAAUUUUCUCAAGGAUAAACAAGGUAUGGGGAUCUAGGCAGACGAGGGAAAAUCAAAUCAAACCCGAAGGCAAAAUUUAAUUGCGUUUAACCUGAAAU ---.((((((................)))))).(((.(((((..(((.(((.(((((.(.....)...)))...))...))).))).....((((.......))))))))).)))..... ( -19.29) >consensus ___GGGGAAAUGCAUAAAAUUUCAAUUUUCUCUAGCAUAAACAAGCCAAAAAGGCUUUGGCAGCCGAGGGAAAAUCAAAUCAAGCCCAGAGGCAAAAUUUAAUUGCGUUUAACCUGAAAU ...((..(((((((..((((((......................((((((.....)))))).(((..(((..............)))...))).))))))...)))))))..))...... (-20.76 = -21.56 + 0.80)

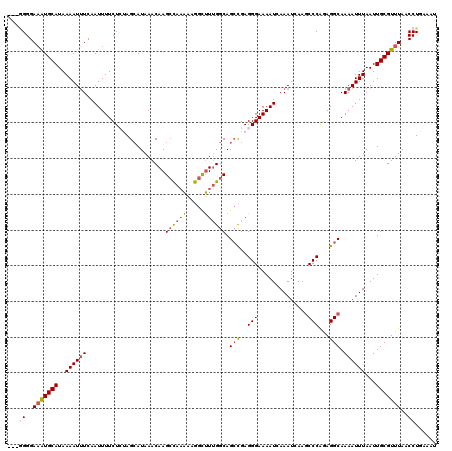
| Location | 15,763,554 – 15,763,674 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.52 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -21.46 |
| Energy contribution | -22.42 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15763554 120 - 23771897 AUUUCAGGUUAAACGCAAUUAAAUUUUGCCUCUGGGCUUGAUUUGAUUUUCCCUCAGCUGCCAAAGCCUUUUUGGCUUGUUUAUGCUAGAGAAAAUUGAAAUUUUAUGCAUUUCCCGCAU ...(((((((....((((.......)))).....)))))))((..((((((.((.(((.....(((((.....)))))......))))).))))))..)).....((((.......)))) ( -31.00) >DroSec_CAF1 15128 117 - 1 AUUUCAGGUUAAACGCAAUUAAAUUUUGCCUCUGGGCUUGAUUUGAUUUUCCCUCGGCUGCCAAAGCCUUUUUGGCUUGUUUAUGCUACAGAAAAUUGAAAUUUUAUGCAUUUCCCC--- ...(((((((....((((.......)))).....)))))))((..((((((....(((.....(((((.....)))))......)))...))))))..)).................--- ( -26.40) >DroSim_CAF1 16123 117 - 1 AUUUCAGGUUAAACGCAAUUAAAUUUUGCCUCUGGGCUUGAUUUCAUUUUCCCUCGGCUGCCAAAGCCUUUUUGGCUUGUUUAUGCUACAGAAAAUUGAAAUUUUAUGCAUUUCCCC--- ......((..(((.(((..........(((....)))..((((((((((((....(((.....(((((.....)))))......)))...))))).)))))))...))).)))..))--- ( -26.30) >DroEre_CAF1 15265 117 - 1 AUUUCAGGUUAAACGCAAUUAAAUUUUGCCUCUGGGCUUGAUUUGAUUUUCCCUCGGCUGCCAAAGCCUUUUUGGCUUGUUUAUGCUGGAGAAAAUUGAAAUUUUAUGCACAUCCCC--- ...(((((((....((((.......)))).....)))))))((..((((((.(.((((.....(((((.....)))))......))))).))))))..)).................--- ( -30.40) >DroAna_CAF1 10382 117 - 1 AUUUCAGGUUAAACGCAAUUAAAUUUUGCCUUCGGGUUUGAUUUGAUUUUCCCUCGUCUGCCUAGAUCCCCAUACCUUGUUUAUCCUUGAGAAAAUUGAGAUUUUAUGCAUUUCCCU--- .(((((((.(((((..(((((((((..(((....)))..))))))))).......((((....))))...........))))).)).))))).....(((((.......)))))...--- ( -23.10) >consensus AUUUCAGGUUAAACGCAAUUAAAUUUUGCCUCUGGGCUUGAUUUGAUUUUCCCUCGGCUGCCAAAGCCUUUUUGGCUUGUUUAUGCUAGAGAAAAUUGAAAUUUUAUGCAUUUCCCC___ (((((((.........(((((((((..(((....)))..)))))))))....((((((.....(((((.....)))))......))).)))....))))))).................. (-21.46 = -22.42 + 0.96)

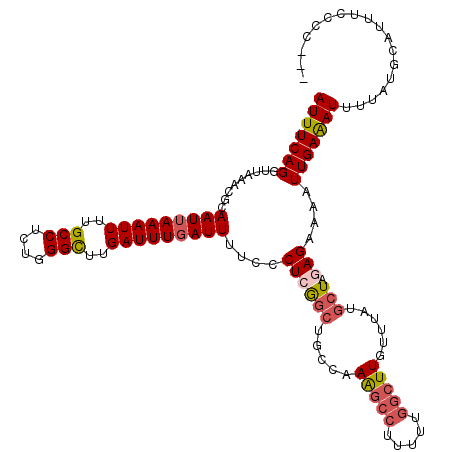
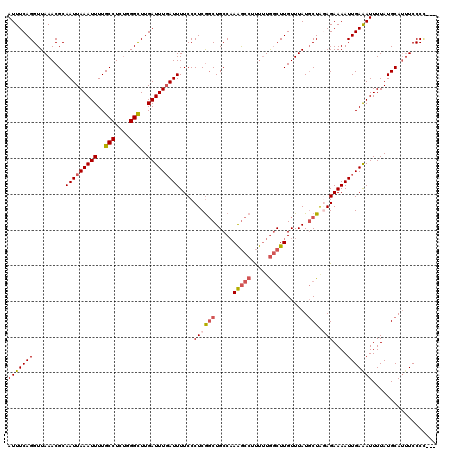
| Location | 15,763,594 – 15,763,714 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -23.90 |
| Energy contribution | -23.54 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15763594 120 + 23771897 ACAAGCCAAAAAGGCUUUGGCAGCUGAGGGAAAAUCAAAUCAAGCCCAGAGGCAAAAUUUAAUUGCGUUUAACCUGAAAUCAAAUUUUCAAGACGAUGCUGACCAGGCCUAGAUUCAAUC ...........((((((.(((((((((..(.....)...))).(((....)))............(((((....(((((......))))))))))..)))).)))))))).......... ( -26.80) >DroSec_CAF1 15165 120 + 1 ACAAGCCAAAAAGGCUUUGGCAGCCGAGGGAAAAUCAAAUCAAGCCCAGAGGCAAAAUUUAAUUGCGUUUAACCUGAAAUCAAAUUUUCAAGACGAUGCUGACCAGGCCUAGAUUCAAUC ...........((((((.((((((((.(((..............)))(((.((((.......)))).)))..(.(((((......))))).).))..)))).)))))))).......... ( -26.64) >DroSim_CAF1 16160 120 + 1 ACAAGCCAAAAAGGCUUUGGCAGCCGAGGGAAAAUGAAAUCAAGCCCAGAGGCAAAAUUUAAUUGCGUUUAACCUGAAAUCAAAUUUUCAAGACGAUGCUGACCAGGCCUAGAUUCAAUC ...........((((((.((((((((.(((....((....))..)))(((.((((.......)))).)))..(.(((((......))))).).))..)))).)))))))).......... ( -28.20) >DroEre_CAF1 15302 120 + 1 ACAAGCCAAAAAGGCUUUGGCAGCCGAGGGAAAAUCAAAUCAAGCCCAGAGGCAAAAUUUAAUUGCGUUUAACCUGAAAUCAAAUUUUCAAGACGAUGCUGACCAGGCCUAGAUUCAAUC ...........((((((.((((((((.(((..............)))(((.((((.......)))).)))..(.(((((......))))).).))..)))).)))))))).......... ( -26.64) >DroAna_CAF1 10419 120 + 1 ACAAGGUAUGGGGAUCUAGGCAGACGAGGGAAAAUCAAAUCAAACCCGAAGGCAAAAUUUAAUUGCGUUUAACCUGAAAUCAAAUUUUGAGGACGAUGCUGACCAGGCCUAGAUUCAAUC ...........((((((((((((((..(((..............)))....((((.......))))))))..(((.(((......))).)))..............)))))))))).... ( -29.44) >consensus ACAAGCCAAAAAGGCUUUGGCAGCCGAGGGAAAAUCAAAUCAAGCCCAGAGGCAAAAUUUAAUUGCGUUUAACCUGAAAUCAAAUUUUCAAGACGAUGCUGACCAGGCCUAGAUUCAAUC ...........((((((.((((((((.(((..............)))(((.((((.......)))).)))..(.(((((......))))).).))..)))).)))))))).......... (-23.90 = -23.54 + -0.36)

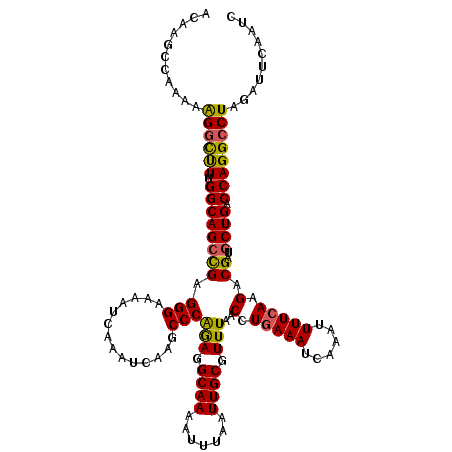
| Location | 15,763,594 – 15,763,714 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -31.84 |
| Consensus MFE | -27.78 |
| Energy contribution | -28.70 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15763594 120 - 23771897 GAUUGAAUCUAGGCCUGGUCAGCAUCGUCUUGAAAAUUUGAUUUCAGGUUAAACGCAAUUAAAUUUUGCCUCUGGGCUUGAUUUGAUUUUCCCUCAGCUGCCAAAGCCUUUUUGGCUUGU ..(..((...((((.(((.((((..((((((((((......)))))))....))).(((((((((..(((....)))..)))))))))........)))))))..)))).))..)..... ( -32.90) >DroSec_CAF1 15165 120 - 1 GAUUGAAUCUAGGCCUGGUCAGCAUCGUCUUGAAAAUUUGAUUUCAGGUUAAACGCAAUUAAAUUUUGCCUCUGGGCUUGAUUUGAUUUUCCCUCGGCUGCCAAAGCCUUUUUGGCUUGU ...........(((..((((.((.....(((((((......)))))))......))(((((((((..(((....)))..))))))))).......))))))).(((((.....))))).. ( -33.20) >DroSim_CAF1 16160 120 - 1 GAUUGAAUCUAGGCCUGGUCAGCAUCGUCUUGAAAAUUUGAUUUCAGGUUAAACGCAAUUAAAUUUUGCCUCUGGGCUUGAUUUCAUUUUCCCUCGGCUGCCAAAGCCUUUUUGGCUUGU ..(..((...((((.(((.((((..((....(((((..(((..(((((((....((((.......)))).....)))))))..))))))))...)))))))))..)))).))..)..... ( -31.80) >DroEre_CAF1 15302 120 - 1 GAUUGAAUCUAGGCCUGGUCAGCAUCGUCUUGAAAAUUUGAUUUCAGGUUAAACGCAAUUAAAUUUUGCCUCUGGGCUUGAUUUGAUUUUCCCUCGGCUGCCAAAGCCUUUUUGGCUUGU ...........(((..((((.((.....(((((((......)))))))......))(((((((((..(((....)))..))))))))).......))))))).(((((.....))))).. ( -33.20) >DroAna_CAF1 10419 120 - 1 GAUUGAAUCUAGGCCUGGUCAGCAUCGUCCUCAAAAUUUGAUUUCAGGUUAAACGCAAUUAAAUUUUGCCUUCGGGUUUGAUUUGAUUUUCCCUCGUCUGCCUAGAUCCCCAUACCUUGU ......((((((((..((.(.((.....(((.(((......))).)))......))(((((((((..(((....)))..))))))))).......).))))))))))............. ( -28.10) >consensus GAUUGAAUCUAGGCCUGGUCAGCAUCGUCUUGAAAAUUUGAUUUCAGGUUAAACGCAAUUAAAUUUUGCCUCUGGGCUUGAUUUGAUUUUCCCUCGGCUGCCAAAGCCUUUUUGGCUUGU ...........(((..((((.((.....(((((((......)))))))......))(((((((((..(((....)))..))))))))).......))))))).(((((.....))))).. (-27.78 = -28.70 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:46 2006