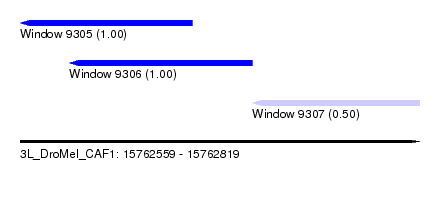
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,762,559 – 15,762,819 |
| Length | 260 |
| Max. P | 0.999963 |

| Location | 15,762,559 – 15,762,671 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.10 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -30.60 |
| Energy contribution | -30.64 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.93 |
| SVM RNA-class probability | 0.999963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15762559 112 - 23771897 CAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUUUGCCACAGAGGCAAACAGAAAACGGCGCUAAAGUUGCUAAAGAAAGUGAACAAACAGAG-------- ....(((((((((..((((((..(((((((((((((....))))).))))))))(((.....))).))))))..)))))......((((((......))).)))...)))).-------- ( -33.00) >DroSec_CAF1 14154 112 - 1 CAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUUUGCCACAGAGGCAAACAGAAAACGGCGCUAAAGUUGCAAAAGAAAGUGAACAAACAGAG-------- ....(((((((((..((((((..(((((((((((((....))))).))))))))(((.....))).))))))..)))))((.......)).................)))).-------- ( -32.60) >DroSim_CAF1 15118 112 - 1 CAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUUUGCCACAGAGGCAAACAGAAAACGGCGCUAAAGUUGCAAAAGAAAGUGAACAAACAGAG-------- ....(((((((((..((((((..(((((((((((((....))))).))))))))(((.....))).))))))..)))))((.......)).................)))).-------- ( -32.60) >DroEre_CAF1 14277 112 - 1 CAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUUUGCCACAGAGGCAAACAGAAAACGGCGCUAAAGUUGCAAAAGAAAGUGAACAAACAGAG-------- ....(((((((((..((((((..(((((((((((((....))))).))))))))(((.....))).))))))..)))))((.......)).................)))).-------- ( -32.60) >DroAna_CAF1 9532 118 - 1 CAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUUUGCCACAGCAGCAAACAGAAAAUGGCGCUAAAGUUGCAAAAGAAAAC--ACAAACACAGACAUACUA ....(((((((((....(((..((((((((((((((....))))).)))))))))..))))))))..............((.......))..........--.......))))....... ( -27.60) >consensus CAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUUUGCCACAGAGGCAAACAGAAAACGGCGCUAAAGUUGCAAAAGAAAGUGAACAAACAGAG________ ....(((((((((..((((((..(((((((((((((....))))).))))))))(((.....))).))))))..)))))((.......)).............))))............. (-30.60 = -30.64 + 0.04)

| Location | 15,762,591 – 15,762,710 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.64 |
| Mean single sequence MFE | -35.57 |
| Consensus MFE | -30.68 |
| Energy contribution | -31.88 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15762591 119 - 23771897 CCGGCUG-UGAGAGCCCAGCACAUAAAAUUCAAUAAAACGCAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUUUGCCACAGAGGCAAACAGAAAACGGCG (((((((-........))))........................((((..((((.(((((..((((((((((((((....))))).)))))))))..)))))))))..))))...))).. ( -36.30) >DroSec_CAF1 14186 119 - 1 CUGGCUG-AGAGAGCCCAGCACAUAAAAUUCAAUAAAACGCAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUUUGCCACAGAGGCAAACAGAAAACGGCG .((((((-........)))).))...............(((...((((..((((.(((((..((((((((((((((....))))).)))))))))..)))))))))..)))).....))) ( -36.40) >DroSim_CAF1 15150 119 - 1 CCGUCUG-AGAGAGCCCAGCACAUAAAAUUCAAUAAAACGCAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUUUGCCACAGAGGCAAACAGAAAACGGCG ..(((((-........))).))................(((...((((..((((.(((((..((((((((((((((....))))).)))))))))..)))))))))..)))).....))) ( -34.00) >DroEre_CAF1 14309 119 - 1 CCGGCUG-AAAGAGCCGAGCACAUAAAAUUCAAUAAAACGCAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUUUGCCACAGAGGCAAACAGAAAACGGCG .(((((.-....))))).....................(((...((((..((((.(((((..((((((((((((((....))))).)))))))))..)))))))))..)))).....))) ( -38.80) >DroAna_CAF1 9570 118 - 1 --AGCAGUGGAGAGACGGGCACAUAAAAUUCAAUAAAACGCAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUUUGCCACAGCAGCAAACAGAAAAUGGCG --.((.((((...(((((.((.(((.................))).)).)))))........((((((((((((((....))))).))))))))))))).)).................. ( -32.33) >consensus CCGGCUG_AGAGAGCCCAGCACAUAAAAUUCAAUAAAACGCAUAUUUGGCUGUCAUUUGUUCCGAGUUUUGGGCGCCAAAGUGCCACAAAAUUUGCCACAGAGGCAAACAGAAAACGGCG ..((((......))))......................(((...((((..((((.(((((..((((((((((((((....))))).)))))))))..)))))))))..)))).....))) (-30.68 = -31.88 + 1.20)



| Location | 15,762,710 – 15,762,819 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 90.77 |
| Mean single sequence MFE | -28.11 |
| Consensus MFE | -25.08 |
| Energy contribution | -24.70 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15762710 109 - 23771897 AAGGAAAUCAAAAGCCGGCUAAGCGCCAAAAAAGCUGUAAGGAAAUAAUGUCUGGCCAGGCCAACGAG------CAAGGAGCGUUAUCCUUGGAGAGGUACGGUGCGAAAAAGCG .................(((..(((((.......((.((((((..((((((....((..((......)------)..)).)))))))))))).))......))))).....))). ( -27.32) >DroSec_CAF1 14305 109 - 1 AAGGAAAUCAAAAGCCGGCUAAGCGCCAAAAAAGCUGUAAGGAAAUAAUGUCUGGCCAAGCCAACGAG------CAAGGAGCGUUAUCCUUGGAGAGGCACCGUGCGAAAAAGCG .............((((((.....))).......((.((((((..((((((....((..((......)------)..)).)))))))))))).)).))).....((......)). ( -27.60) >DroSim_CAF1 15269 109 - 1 AAGGAAAUCAAAAGCCGGCUAAGCGCCAAAAAAGCUGUAAGGAAAUAAUGUCUGGCCAGGCCAACGAG------CAAGGAGCGUUAUCCUUGGAGAGGCACCGUGCGAAAAAGCG .............((((((.....))).......((.((((((..((((((....((..((......)------)..)).)))))))))))).)).))).....((......)). ( -28.60) >DroEre_CAF1 14428 105 - 1 ----------AAAGCCGGCUAAGCGCCAAAAAAGCUGUAAGGAAAUAAUGUCUGGCCAAGUCAACGAGCGGCAUCAAGGAGCGUUAUCCUUGGAGAGGCACGGUGCGAAAAAGCG ----------...((((((.....))).......((.((((((..((((((((.(((............))).....))).))))))))))).)).))).....((......)). ( -28.90) >consensus AAGGAAAUCAAAAGCCGGCUAAGCGCCAAAAAAGCUGUAAGGAAAUAAUGUCUGGCCAAGCCAACGAG______CAAGGAGCGUUAUCCUUGGAGAGGCACCGUGCGAAAAAGCG .............((((((.....))).......((.((((((..(((((((((((...)))...(........)..))).))))))))))).)).))).....((......)). (-25.08 = -24.70 + -0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:41 2006