
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,757,519 – 15,757,826 |
| Length | 307 |
| Max. P | 0.977479 |

| Location | 15,757,519 – 15,757,628 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.49 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -13.16 |
| Energy contribution | -16.74 |
| Covariance contribution | 3.58 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977479 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15757519 109 + 23771897 U------UAAAACUUAUCCUUAUCACACAACCGCAACUAAACGAUUGUAACACGAUCAGUGCUAUAAUUUAGUUGCUGACUUGGCCUUCAGCUUU-----GGGCUCAUGGGUUUGGCCUG .------..................((.(((((((((((...(((((((.(((.....))).))))))))))))))...(.((((((........-----)))).)).)))))))..... ( -26.90) >DroSec_CAF1 9210 109 + 1 U------UAAAACAUAUCCUUAUCACCCAACCGCAACUAAACGAUUGUAACACGAUCAGUGCUAUAAUUUAGUUGCUGACUUGGCCUUUAGCUUU-----GGUCACAUGGGUUGGGCCUG .------..................((((((((((((((...(((((((.(((.....))).))))))))))))))(((((.(((.....)))..-----)))))....))))))).... ( -36.30) >DroSim_CAF1 9445 109 + 1 U------UAAAACAUAUCCUUAUCACCCAACCGCAACUAAACGAUUGUAACACGAUCAGUGCUAUAAUUUAGUUGCUGACUUGGCCUUUAGCUUU-----GGGCACAUGGGUUGGGCCUG .------..................((((((((((((((...(((((((.(((.....))).))))))))))))))...(.((((((........-----)))).)).)))))))).... ( -35.50) >DroEre_CAF1 9333 108 + 1 U------UAU-CCUUAUCCUUAUCACCCAACGGCAACUAAACGAUUGUAACACGAUCAGUGCCAUCAUUUAGUUGCUGGCUUGGCCUUUAGCUUU-----GGGGACAUGGGUUGGGCAUG .------...-(((.((((...((.((((((((((((((((.(((.(...(((.....)))).))).)))))))))))(((........))).))-----)))))...)))).))).... ( -35.50) >DroAna_CAF1 3953 103 + 1 UAGCAAAUAUCAUUUAUUCUUAUCGUGAA-----AACUAAACGUUUGUAACGCGAUCAUUACUAUCGUUUAG------GCCAGGCCCUGAAAAAUACCCUCCCCAAAC-UGUUG-----G (((((................((((((.(-----(((.....))))....))))))......(((..(((((------(......))))))..)))............-)))))-----. ( -14.10) >consensus U______UAAAACUUAUCCUUAUCACCCAACCGCAACUAAACGAUUGUAACACGAUCAGUGCUAUAAUUUAGUUGCUGACUUGGCCUUUAGCUUU_____GGGCACAUGGGUUGGGCCUG .........................((((((((((((((((...(((((.(((.....))).))))))))))))))......(((.....)))................))))))).... (-13.16 = -16.74 + 3.58)



| Location | 15,757,519 – 15,757,628 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.49 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -18.08 |
| Energy contribution | -20.46 |
| Covariance contribution | 2.38 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15757519 109 - 23771897 CAGGCCAAACCCAUGAGCCC-----AAAGCUGAAGGCCAAGUCAGCAACUAAAUUAUAGCACUGAUCGUGUUACAAUCGUUUAGUUGCGGUUGUGUGAUAAGGAUAAGUUUUA------A ..(((((......)).))).-----((((((.....((..((((((((((((((..((((((.....)))))).....)))))))))).......))))..))...)))))).------. ( -26.61) >DroSec_CAF1 9210 109 - 1 CAGGCCCAACCCAUGUGACC-----AAAGCUAAAGGCCAAGUCAGCAACUAAAUUAUAGCACUGAUCGUGUUACAAUCGUUUAGUUGCGGUUGGGUGAUAAGGAUAUGUUUUA------A ...((((((((....((((.-----...((.....))...))))((((((((((..((((((.....)))))).....)))))))))))))))))).................------. ( -35.30) >DroSim_CAF1 9445 109 - 1 CAGGCCCAACCCAUGUGCCC-----AAAGCUAAAGGCCAAGUCAGCAACUAAAUUAUAGCACUGAUCGUGUUACAAUCGUUUAGUUGCGGUUGGGUGAUAAGGAUAUGUUUUA------A ...((((((((..((.(((.-----.........))))).....((((((((((..((((((.....)))))).....)))))))))))))))))).................------. ( -34.60) >DroEre_CAF1 9333 108 - 1 CAUGCCCAACCCAUGUCCCC-----AAAGCUAAAGGCCAAGCCAGCAACUAAAUGAUGGCACUGAUCGUGUUACAAUCGUUUAGUUGCCGUUGGGUGAUAAGGAUAAGG-AUA------A ....((....((.(((((((-----((.......(((...))).(((((((((((((.((((.....))))....)))))))))))))..))))).)))).))....))-...------. ( -37.30) >DroAna_CAF1 3953 103 - 1 C-----CAACA-GUUUGGGGAGGGUAUUUUUCAGGGCCUGGC------CUAAACGAUAGUAAUGAUCGCGUUACAAACGUUUAGUU-----UUCACGAUAAGAAUAAAUGAUAUUUGCUA .-----.....-(((((((..((((..........))))..)------))))))....((((((....))))))...((((((.((-----((......)))).)))))).......... ( -21.60) >consensus CAGGCCCAACCCAUGUGCCC_____AAAGCUAAAGGCCAAGUCAGCAACUAAAUUAUAGCACUGAUCGUGUUACAAUCGUUUAGUUGCGGUUGGGUGAUAAGGAUAAGUUUUA______A ...((((((((.......................(((...))).((((((((((..((((((.....)))))).....))))))))))))))))))........................ (-18.08 = -20.46 + 2.38)



| Location | 15,757,593 – 15,757,706 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.54 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -24.76 |
| Energy contribution | -25.04 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15757593 113 - 23771897 CCUUU--UGCGGCACAUGUGUUUAUGAAUACAUAGAAAUUUUUGACAUUUUUAUGGCUGCUCUGGGCAUUUACUGUAGUCCAGGCCAAACCCAUGAGCCC-----AAAGCUGAAGGCCAA (((((--.(((((.((((.((((.......(((((((((.......)))))))))...((.((((((..........)))))))).)))).)))).))).-----...)).))))).... ( -33.20) >DroSec_CAF1 9284 113 - 1 CCUUU--UGCGGCACAUGUGUUUAUGAAUACAUAGAAAUUUUUGACAUUUUUAUGGCUGCUCUGGGCAUUUACCGUGGCCCAGGCCCAACCCAUGUGACC-----AAAGCUAAAGGCCAA (((((--.((((((((((.(((........(((((((((.......)))))))))...((.((((((..........))))))))..))).)))))).))-----...)).))))).... ( -35.20) >DroSim_CAF1 9519 113 - 1 CCUUU--UGCGGCACAUGUGUUUAUGAAUACAUAGAAAUUUUUGACAUUUUUAUGGCUGCUCUGGGCAUUUACUGAGGCCCAGGCCCAACCCAUGUGCCC-----AAAGCUAAAGGCCAA (((((--.((((((((((.(((........(((((((((.......)))))))))...((.((((((.((....)).))))))))..))).)))))))).-----...)).))))).... ( -39.10) >DroEre_CAF1 9406 113 - 1 CCUUU--UGCGGCACAUGUGUUUAUGAAUACAUAGAAAUUUUUGACAUUUUUAUGGCUGCUCUGGGCAUUUACUGGGGCCCAUGCCCAACCCAUGUCCCC-----AAAGCUAAAGGCCAA (((((--.(((((.((((....))))....(((((((((.......))))))))))))))...((((((......((((....)))).....))))))..-----......))))).... ( -31.80) >DroAna_CAF1 4022 114 - 1 UCCUUUGUGCAACACAUGUGUUUAUGAAUACAUAGAAAUUUUUGACAUUUUUAUGGCUGCCCUGGGCAUUUAUUUUUGCCC-----CAACA-GUUUGGGGAGGGUAUUUUUCAGGGCCUG ...((..((.((((....))))))..))..(((((((((.......)))))))))...((((((((.(..(((((...(((-----(((..-..)))))).)))))..)))))))))... ( -31.30) >consensus CCUUU__UGCGGCACAUGUGUUUAUGAAUACAUAGAAAUUUUUGACAUUUUUAUGGCUGCUCUGGGCAUUUACUGUGGCCCAGGCCCAACCCAUGUGCCC_____AAAGCUAAAGGCCAA (((((...(((((.((((....))))....(((((((((.......))))))))))))))(((((((..........)))))))...........................))))).... (-24.76 = -25.04 + 0.28)


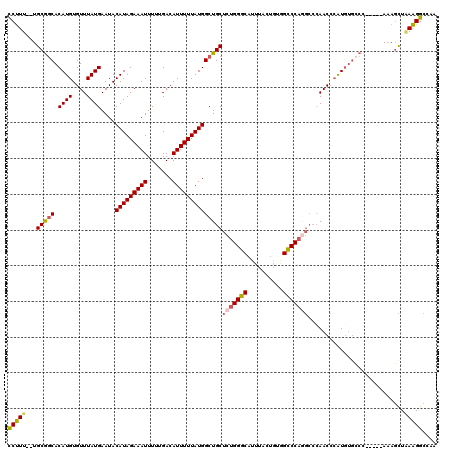
| Location | 15,757,628 – 15,757,746 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.85 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -26.40 |
| Energy contribution | -26.64 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15757628 118 - 23771897 UUUCUCCUUGGGGUCGCUUGCCAUCGACUUUGAGUUGCCUCCUUU--UGCGGCACAUGUGUUUAUGAAUACAUAGAAAUUUUUGACAUUUUUAUGGCUGCUCUGGGCAUUUACUGUAGUC ......((..((((((........))))))..)).(((((.....--.(((((.((((....))))....(((((((((.......))))))))))))))...)))))............ ( -31.70) >DroSec_CAF1 9319 118 - 1 UUUCUCCUUGGGGUCGCUUGCCAUCGACUUUGAGUUGCCUCCUUU--UGCGGCACAUGUGUUUAUGAAUACAUAGAAAUUUUUGACAUUUUUAUGGCUGCUCUGGGCAUUUACCGUGGCC ......((..((((((........))))))..)).(((((.....--.(((((.((((....))))....(((((((((.......))))))))))))))...)))))............ ( -31.70) >DroSim_CAF1 9554 118 - 1 UUUCUCCUUGGGGUCGCUUGCCAUCGACUGUGAGUUGCCUCCUUU--UGCGGCACAUGUGUUUAUGAAUACAUAGAAAUUUUUGACAUUUUUAUGGCUGCUCUGGGCAUUUACUGAGGCC .....(((..((((.(((..(........)..))).))).(((..--.(((((.((((....))))....(((((((((.......))))))))))))))...)))......)..))).. ( -34.20) >DroEre_CAF1 9441 118 - 1 UUUCUCCUUGGGGUCGCUUGGCAUCGACUUUGAGCUGCCUCCUUU--UGCGGCACAUGUGUUUAUGAAUACAUAGAAAUUUUUGACAUUUUUAUGGCUGCUCUGGGCAUUUACUGGGGCC .....(((..((((.(((..(........)..))).))).(((..--.(((((.((((....))))....(((((((((.......))))))))))))))...)))......)..))).. ( -34.10) >DroAna_CAF1 4056 118 - 1 UUUCUCCUUGGGGUCGCUUGCCAUCGACUUGGA--CGCCUUCCUUUGUGCAACACAUGUGUUUAUGAAUACAUAGAAAUUUUUGACAUUUUUAUGGCUGCCCUGGGCAUUUAUUUUUGCC .........(((((.((((((...(((...(((--.....))).))).))))..((((....))))....(((((((((.......))))))))))).))))).((((........)))) ( -25.80) >consensus UUUCUCCUUGGGGUCGCUUGCCAUCGACUUUGAGUUGCCUCCUUU__UGCGGCACAUGUGUUUAUGAAUACAUAGAAAUUUUUGACAUUUUUAUGGCUGCUCUGGGCAUUUACUGUGGCC ......(((..(((((........)))))..)))..((((........(((((.((((....))))....(((((((((.......))))))))))))))...))))............. (-26.40 = -26.64 + 0.24)

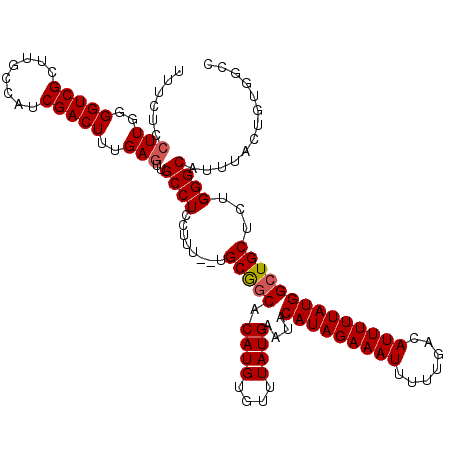
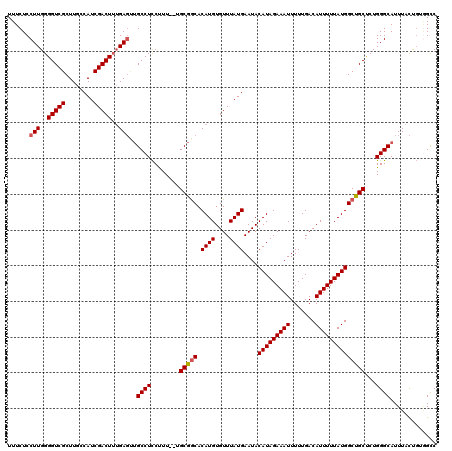
| Location | 15,757,706 – 15,757,826 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -26.39 |
| Consensus MFE | -22.20 |
| Energy contribution | -22.95 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15757706 120 + 23771897 AGGCAACUCAAAGUCGAUGGCAAGCGACCCCAAGGAGAAACCCAAAACCCCAAAAACACGCCAACGCAUUUCGACCACAAAGUUGUCGAGGACACUCCGCACUUCGCUCCCCAUAUAUGU .(((........))).((((..(((((......((((......................((....)).(((((((.((...)).)))))))...)))).....)))))..))))...... ( -27.10) >DroSec_CAF1 9397 120 + 1 AGGCAACUCAAAGUCGAUGGCAAGCGACCCCAAGGAGAAACCCAAAACCCCAAAAACACGCCAACGCAUUUCGACCACAAAGUUGUCGAGGACACUCCGCACUUCGCUCCCCAUAUAUGU .(((........))).((((..(((((......((((......................((....)).(((((((.((...)).)))))))...)))).....)))))..))))...... ( -27.10) >DroSim_CAF1 9632 120 + 1 AGGCAACUCACAGUCGAUGGCAAGCGACCCCAAGGAGAAACCCAAAACCCCAAAAACACGCCAACGCAUUUCGACCACAAAGUUGUCGAGGACACUCCGCACUUCGCUCCCCAUAUAUGU ((....)).(((....((((..(((((......((((......................((....)).(((((((.((...)).)))))))...)))).....)))))..))))...))) ( -27.90) >DroEre_CAF1 9519 117 + 1 AGGCAGCUCAAAGUCGAUGCCAAGCGACCCCAAGGAGAAACACAAAACCCCCA-AACACGCCAACGCAAUUCGACCACAAAGUUGUCGAGGACACUCCACACUUCAGUCCACAUAUAU-- .(((((((....((((((((...(((.......((.............))...-....)))....)))..))))).....)))))))..((((.............))))........-- ( -23.48) >consensus AGGCAACUCAAAGUCGAUGGCAAGCGACCCCAAGGAGAAACCCAAAACCCCAAAAACACGCCAACGCAUUUCGACCACAAAGUUGUCGAGGACACUCCGCACUUCGCUCCCCAUAUAUGU .(((........))).((((..(((((......((((......................((....))..((((((.((...)).))))))....)))).....)))))..))))...... (-22.20 = -22.95 + 0.75)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:31 2006