
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,755,834 – 15,756,102 |
| Length | 268 |
| Max. P | 0.897396 |

| Location | 15,755,834 – 15,755,946 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -21.02 |
| Energy contribution | -21.96 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15755834 112 + 23771897 AAUGUCGAAAGCAAUUUCCUGGAUGGGUUCGUAGAGGAAAAUUGCCGGCGGAACAAGGCAAUUCAAGAUUCAAAACAUAUAGAAUUGGGCUAAUGCCUUUGGAUUUUUAGAU ..(((((...((((((((((..(((....)))..))).))))))))))))..............((((((((((.((........))(((....)))))))))))))..... ( -25.80) >DroSec_CAF1 7477 110 + 1 AAUGUCGAAAGCAAUUUCCUGGAUGGGUUCAUAGAGGAAAAUUGCCGGCGGAACAAGGCAAUUGAAGAUUCAAAG--UAUAGAACUGGGCUAAUGCCCUUAGAUUUUUAGAU ..(((((...((((((((((..(((....)))..))).))))))))))))...........((((((((..((((--(.....)))((((....))))))..)))))))).. ( -27.60) >DroSim_CAF1 7742 102 + 1 AAUGUCGAAAGCAAUUUCCUGGA--------UAGAGGAAAAUUGCCGGCGGAACAAGGCAAUUGAAGAUUCAAAG--UAUAGAACUGGGCUAAUGCCUCUGAAUUUUCAGAU ..(((((...((((((((((...--------...))).))))))))))))...........(((((((((((.((--(.....)))((((....)))).))))))))))).. ( -29.30) >DroEre_CAF1 7580 110 + 1 AAUGUCGAAAGCAAUUUCCUGGACGGGUUCAUGGGGGCAAAUUGCAGUCGGAACAAGGCAAUUGAAGAUUCAAAG--UGCAGGACUGGGACAAUGCUUUCAGAUUUUCAGAU ...((((((((((.(.(((..(.(..((((.(((..((.....))..)))))))..)(((.((((....))))..--)))....)..))).).))))))).)))........ ( -30.20) >consensus AAUGUCGAAAGCAAUUUCCUGGAUGGGUUCAUAGAGGAAAAUUGCCGGCGGAACAAGGCAAUUGAAGAUUCAAAG__UAUAGAACUGGGCUAAUGCCUUUAGAUUUUCAGAU ..((((....((((((((((..(((....)))..))).))))))).(......)..)))).((((((((((((.............((((....)))))))))))))))).. (-21.02 = -21.96 + 0.94)

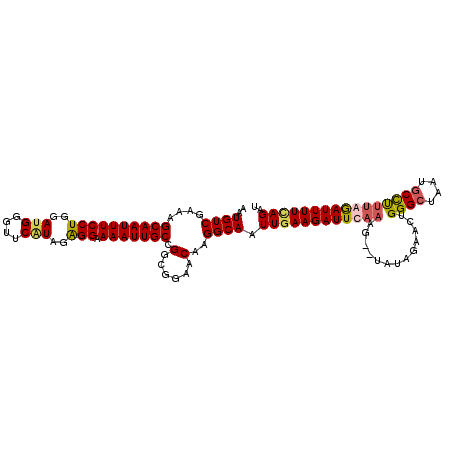

| Location | 15,755,834 – 15,755,946 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 85.59 |
| Mean single sequence MFE | -21.36 |
| Consensus MFE | -15.59 |
| Energy contribution | -16.34 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15755834 112 - 23771897 AUCUAAAAAUCCAAAGGCAUUAGCCCAAUUCUAUAUGUUUUGAAUCUUGAAUUGCCUUGUUCCGCCGGCAAUUUUCCUCUACGAACCCAUCCAGGAAAUUGCUUUCGACAUU .............((((((........((((..........)))).......))))))....((..((((((((.(((..............)))))))))))..))..... ( -18.60) >DroSec_CAF1 7477 110 - 1 AUCUAAAAAUCUAAGGGCAUUAGCCCAGUUCUAUA--CUUUGAAUCUUCAAUUGCCUUGUUCCGCCGGCAAUUUUCCUCUAUGAACCCAUCCAGGAAAUUGCUUUCGACAUU .............((((((........((((....--....)))).......))))))....((..((((((((.(((..(((....)))..)))))))))))..))..... ( -22.86) >DroSim_CAF1 7742 102 - 1 AUCUGAAAAUUCAGAGGCAUUAGCCCAGUUCUAUA--CUUUGAAUCUUCAAUUGCCUUGUUCCGCCGGCAAUUUUCCUCUA--------UCCAGGAAAUUGCUUUCGACAUU .(((((....)))))((((........((((....--....)))).......))))......((..((((((((.(((...--------...)))))))))))..))..... ( -23.46) >DroEre_CAF1 7580 110 - 1 AUCUGAAAAUCUGAAAGCAUUGUCCCAGUCCUGCA--CUUUGAAUCUUCAAUUGCCUUGUUCCGACUGCAAUUUGCCCCCAUGAACCCGUCCAGGAAAUUGCUUUCGACAUU ...........((((((((...(((.......(((--..((((....)))).)))...((((.....((.....))......)))).......)))...))))))))..... ( -20.50) >consensus AUCUAAAAAUCCAAAGGCAUUAGCCCAGUUCUAUA__CUUUGAAUCUUCAAUUGCCUUGUUCCGCCGGCAAUUUUCCUCUAUGAACCCAUCCAGGAAAUUGCUUUCGACAUU .............((((((........((((..........)))).......))))))....((..((((((((.(((..(((....)))..)))))))))))..))..... (-15.59 = -16.34 + 0.75)


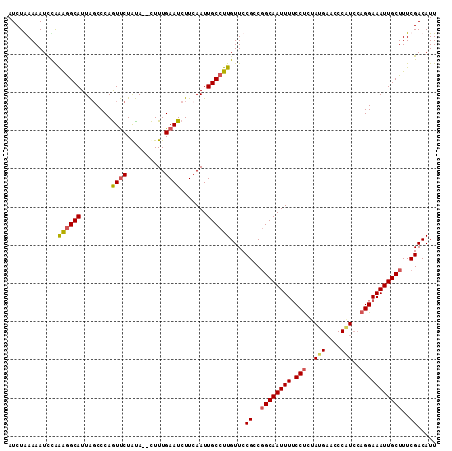
| Location | 15,755,871 – 15,755,986 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 88.45 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -21.61 |
| Energy contribution | -22.73 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15755871 115 + 23771897 AAAAUUGCCGGCGGAACAAGGCAAUUCAAGAUUCAAAACAUAUAGAAUUGGGCUAAUGCCUUUGGAUUUUUAGAUGGCUGUUGUUAAGCCCAAUAAAAGCAAAAGCCAAUGCCGU ........(((((......(((.(((((((((((..........)))))((((....))))))))))(((((...((((.......))))...)))))......)))..))))). ( -27.60) >DroSec_CAF1 7514 113 + 1 AAAAUUGCCGGCGGAACAAGGCAAUUGAAGAUUCAAAG--UAUAGAACUGGGCUAAUGCCCUUAGAUUUUUAGAUGGCUGUUGUUAAGCCCAAUAAAAGCAAAAGCCAAUGCCUU .................((((((.((((((((..((((--(.....)))((((....))))))..)))))))).(((((.(((((............))))).))))).)))))) ( -28.70) >DroSim_CAF1 7771 113 + 1 AAAAUUGCCGGCGGAACAAGGCAAUUGAAGAUUCAAAG--UAUAGAACUGGGCUAAUGCCUCUGAAUUUUCAGAUGACUGUUGUUAAGCCCAAUAAAAGCAAAAGCCAAUGCCAU ...((((..(((..(((((((((.(((((((((((.((--(.....)))((((....)))).))))))))))).)).)).)))))..)))))))....(((........)))... ( -29.60) >DroEre_CAF1 7617 113 + 1 CAAAUUGCAGUCGGAACAAGGCAAUUGAAGAUUCAAAG--UGCAGGACUGGGACAAUGCUUUCAGAUUUUCAGAUGGCUGUUGUUAAGCCCAAUAAAGGCCAAAGCCAUUGCCGU ...........(((..((((((..(((((((((..(((--.(((............))))))..))))))))).(((((.(((((......))))).)))))..))).)))))). ( -26.50) >consensus AAAAUUGCCGGCGGAACAAGGCAAUUGAAGAUUCAAAG__UAUAGAACUGGGCUAAUGCCUUUAGAUUUUCAGAUGGCUGUUGUUAAGCCCAAUAAAAGCAAAAGCCAAUGCCGU ...................((((.((((((((((((.............((((....)))))))))))))))).(((((.(((((............))))).))))).)))).. (-21.61 = -22.73 + 1.12)

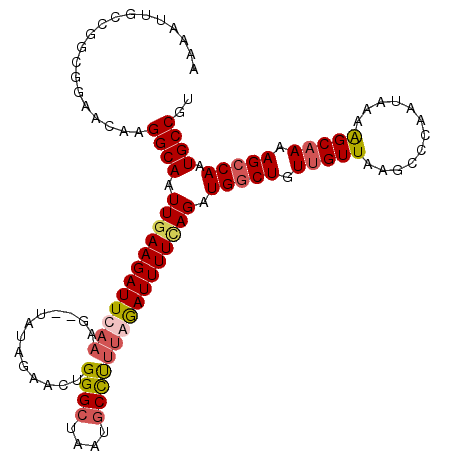
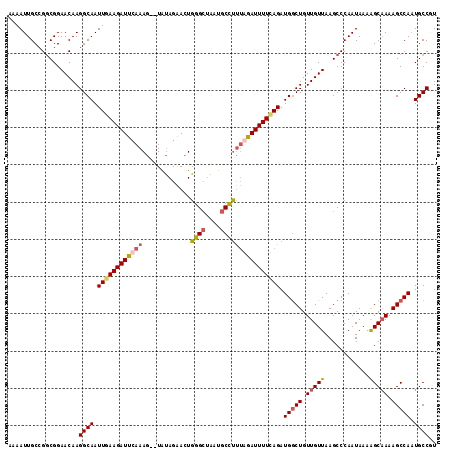
| Location | 15,755,871 – 15,755,986 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 88.45 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -20.75 |
| Energy contribution | -20.50 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15755871 115 - 23771897 ACGGCAUUGGCUUUUGCUUUUAUUGGGCUUAACAACAGCCAUCUAAAAAUCCAAAGGCAUUAGCCCAAUUCUAUAUGUUUUGAAUCUUGAAUUGCCUUGUUCCGCCGGCAAUUUU .((((...((((..((((((((((.((((.......)))).......)))..)))))))..))))((((((.................)))))).........))))........ ( -27.53) >DroSec_CAF1 7514 113 - 1 AAGGCAUUGGCUUUUGCUUUUAUUGGGCUUAACAACAGCCAUCUAAAAAUCUAAGGGCAUUAGCCCAGUUCUAUA--CUUUGAAUCUUCAAUUGCCUUGUUCCGCCGGCAAUUUU ((((((..((((..((((((((...((((.......))))...........))))))))..))))..((((....--....)))).......))))))................. ( -25.94) >DroSim_CAF1 7771 113 - 1 AUGGCAUUGGCUUUUGCUUUUAUUGGGCUUAACAACAGUCAUCUGAAAAUUCAGAGGCAUUAGCCCAGUUCUAUA--CUUUGAAUCUUCAAUUGCCUUGUUCCGCCGGCAAUUUU ((((.(((((((..((((.......((((.......)))).(((((....)))))))))..)).))))).)))).--............(((((((..........))))))).. ( -27.30) >DroEre_CAF1 7617 113 - 1 ACGGCAAUGGCUUUGGCCUUUAUUGGGCUUAACAACAGCCAUCUGAAAAUCUGAAAGCAUUGUCCCAGUCCUGCA--CUUUGAAUCUUCAAUUGCCUUGUUCCGACUGCAAUUUG ..(((((((.(((((((((.....)))))......(((....)))........))))))))))).(((((..(((--..((((....))))......)))...)))))....... ( -24.30) >consensus ACGGCAUUGGCUUUUGCUUUUAUUGGGCUUAACAACAGCCAUCUAAAAAUCCAAAGGCAUUAGCCCAGUUCUAUA__CUUUGAAUCUUCAAUUGCCUUGUUCCGCCGGCAAUUUU ..(((....(((((((.(((((...((((.......))))...)))))...)))))))....)))........................(((((((..........))))))).. (-20.75 = -20.50 + -0.25)



| Location | 15,755,946 – 15,756,062 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.25 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -25.98 |
| Energy contribution | -25.98 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15755946 116 - 23771897 GA----AUAUUAGCCAUACUGAUUGUGAAAUUCUGCAAUUUGCCUGGCUAACUUGUGUGCCGAAAUGCUUUUGUUAAUUGACGGCAUUGGCUUUUGCUUUUAUUGGGCUUAACAACAGCC ..----...(((((((..(.((((((........)))))).)..))))))).((((..(((..((((((.(((.....))).))))))(((....))).......)))...))))..... ( -28.20) >DroSec_CAF1 7587 120 - 1 GUACAUAUAUUAGCUAUACUGAUUGUGAAAUUCUGCAAUUUGCCUGGCCAACUUGUGUGCCGAAAUGCUUUUGUUAAUUGAAGGCAUUGGCUUUUGCUUUUAUUGGGCUUAACAACAGCC (((((((..((.((((..(.((((((........)))))).)..)))).))..)))))))......(((.(((((((..((((((....))))))((((.....))))))))))).))). ( -31.60) >DroSim_CAF1 7844 116 - 1 GA----AUAUUAGCCAUACUGAUUGUAAAAUUCUGCAAUUUGCCUGGCUAACUAGUGUGCCGAAAUGCUUUUGUUAAUUGAUGGCAUUGGCUUUUGCUUUUAUUGGGCUUAACAACAGUC ..----...(((((((..(.(((((((......))))))).)..)))))))((((((.((.((((.(((..(((((.....)))))..)))))))))...)))))).............. ( -28.50) >DroEre_CAF1 7690 116 - 1 UA----AAAUUGGCCACAAUUAUUGUGAAAUUCUGCAGUUUGUCUGGCCAACUUGUGUGCCGAAAUGCUUUUGUUAAUUGACGGCAAUGGCUUUGGCCUUUAUUGGGCUUAACAACAGCC ..----...(((((((.....(((((........))))).....)))))))......(((((.(((((....))..)))..)))))..(((((((((((.....))))....))).)))) ( -30.20) >consensus GA____AUAUUAGCCAUACUGAUUGUGAAAUUCUGCAAUUUGCCUGGCCAACUUGUGUGCCGAAAUGCUUUUGUUAAUUGACGGCAUUGGCUUUUGCUUUUAUUGGGCUUAACAACAGCC .........(((((((....(((((((......)))))))....))))))).((((..(((..((((((.(((.....))).))))))(((....))).......)))...))))..... (-25.98 = -25.98 + -0.00)

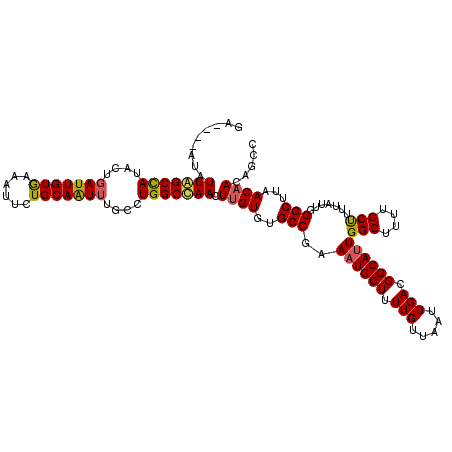
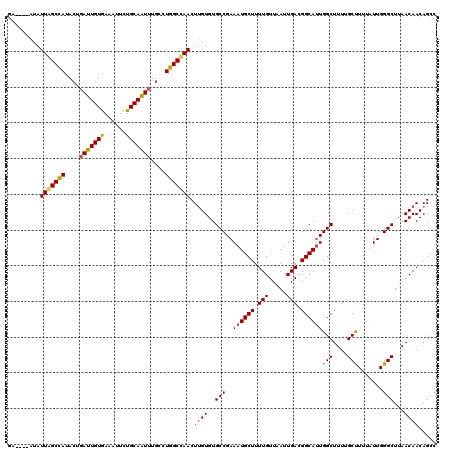
| Location | 15,755,986 – 15,756,102 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.39 |
| Mean single sequence MFE | -18.07 |
| Consensus MFE | -13.26 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15755986 116 + 23771897 CAAUUAACAAAAGCAUUUCGGCACACAAGUUAGCCAGGCAAAUUGCAGAAUUUCACAAUCAGUAUGGCUAAUAUUCCUUAAACAUUCUUUUGUGCGCCUUCUAUAAAUACUAGGUC ...................(((.(((((((((((((.((..((((..........))))..)).)))))))).................))))).))).................. ( -21.82) >DroSim_CAF1 7884 89 + 1 CAAUUAACAAAAGCAUUUCGGCACACUAGUUAGCCAGGCAAAUUGCAGAAUUUUACAAUCAGUAUGGCUAAUAUUCCAUAAA---------------------------CUGGGUC ............((......))......((((((((.((..((((..........))))..)).))))))))..((((....---------------------------.)))).. ( -16.10) >DroEre_CAF1 7730 100 + 1 CAAUUAACAAAAGCAUUUCGGCACACAAGUUGGCCAGACAAACUGCAGAAUUUCACAAUAAUUGUGGCCAAUUUUACCUAAAUAUCGCUAUGA----------------CCGAAAC ...............((((((.....(((((((((((.....)))........((((.....))))))))))))..........((.....))----------------)))))). ( -16.30) >consensus CAAUUAACAAAAGCAUUUCGGCACACAAGUUAGCCAGGCAAAUUGCAGAAUUUCACAAUCAGUAUGGCUAAUAUUCCAUAAA_AU___U_UG_________________CUGGGUC ............((......))......((((((((.((..((((..........))))..)).))))))))............................................ (-13.26 = -13.27 + 0.00)

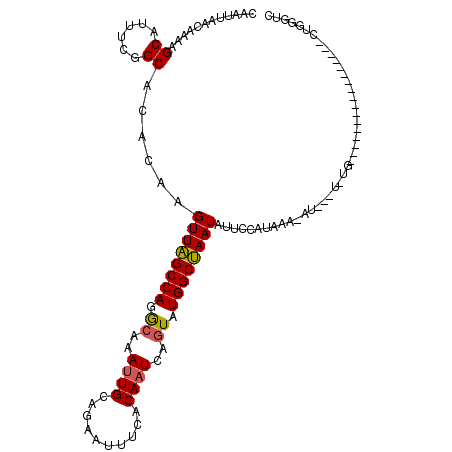
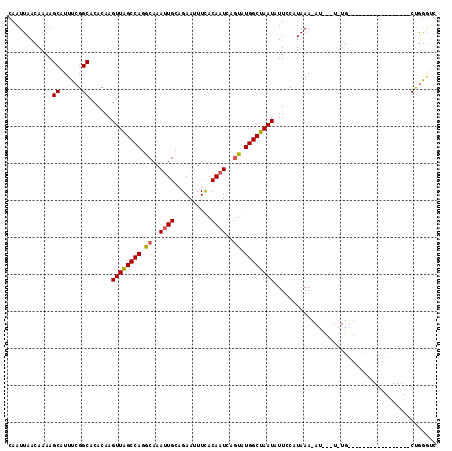
| Location | 15,755,986 – 15,756,102 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.39 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -15.12 |
| Energy contribution | -14.57 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15755986 116 - 23771897 GACCUAGUAUUUAUAGAAGGCGCACAAAAGAAUGUUUAAGGAAUAUUAGCCAUACUGAUUGUGAAAUUCUGCAAUUUGCCUGGCUAACUUGUGUGCCGAAAUGCUUUUGUUAAUUG (((..(((((((......(((((((((...(((((((...)))))))(((((..(.((((((........)))))).)..)))))...))))))))).)))))))...)))..... ( -33.00) >DroSim_CAF1 7884 89 - 1 GACCCAG---------------------------UUUAUGGAAUAUUAGCCAUACUGAUUGUAAAAUUCUGCAAUUUGCCUGGCUAACUAGUGUGCCGAAAUGCUUUUGUUAAUUG ....(((---------------------------((.(..(((..(((((((..(.(((((((......))))))).)..)))))))...((((......)))))))..).))))) ( -19.50) >DroEre_CAF1 7730 100 - 1 GUUUCGG----------------UCAUAGCGAUAUUUAGGUAAAAUUGGCCACAAUUAUUGUGAAAUUCUGCAGUUUGUCUGGCCAACUUGUGUGCCGAAAUGCUUUUGUUAAUUG (((((((----------------((((((...(((....)))...(((((((.....(((((........))))).....))))))).))))).)))))))).............. ( -24.50) >consensus GACCCAG_________________CA_A___AU_UUUAAGGAAUAUUAGCCAUACUGAUUGUGAAAUUCUGCAAUUUGCCUGGCUAACUUGUGUGCCGAAAUGCUUUUGUUAAUUG .............................................(((((((....(((((((......)))))))....)))))))...((((......))))............ (-15.12 = -14.57 + -0.55)

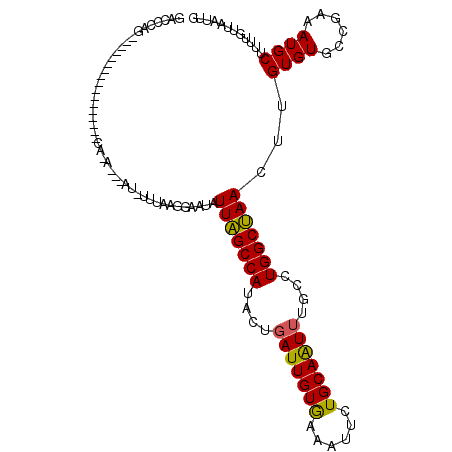
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:25 2006