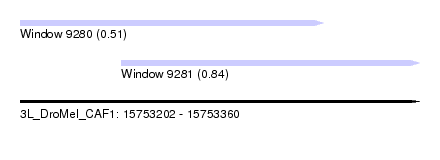
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,753,202 – 15,753,360 |
| Length | 158 |
| Max. P | 0.836113 |

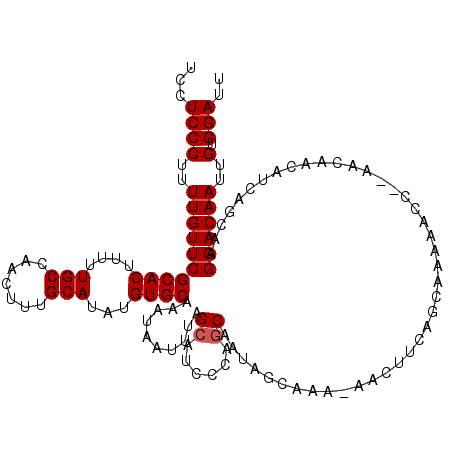
| Location | 15,753,202 – 15,753,322 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -17.76 |
| Consensus MFE | -11.74 |
| Energy contribution | -12.26 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15753202 120 + 23771897 UUGAACCUCUACUUUCAACCGACUAUCCGUUCAUUUUUGCUCCUCCCUUUUGUUCGCACUUUUUGCCAACUUUGCAUAUGUGCAAAUAAUUUAGCAUCCCAGCAAUAGCAAAAAACUUCA .(((((........((....))......)))))((((((((........(((((.((((....(((.......)))...)))).)))))....((......))...))))))))...... ( -19.54) >DroSec_CAF1 4894 119 + 1 UUGAACCUCUACUUUCAACCGACUAUCCGUUCAUUUUUGCUCCUCCCUUUUGUUCGCACUUUUUGCCAACUUUGCAUAUGUGCAAAUAAUUUAGCAUCCCAGCAAUAGCAAA-AACUUCA .(((((........((....))......)))))((((((((........(((((.((((....(((.......)))...)))).)))))....((......))...))))))-))..... ( -19.24) >DroSim_CAF1 5143 119 + 1 UUGAACCUCCACUUUCAACAGACUAUCCGUUCUUUUUUGCUCCUCCCUUUUGUUCGCACUUUUUGCCAACUUUGCAUAUGUGCAAAUAAUUUAGCAUCCCAGCAAUAGCAAA-AACUUCA (((((........)))))...............((((((((........(((((.((((....(((.......)))...)))).)))))....((......))...))))))-))..... ( -18.40) >DroEre_CAF1 4961 119 + 1 UUGAACCUCCACUUUCAACCGACUAUCCGCUCAUUUUUGCUCCUCCCUUUUGUUCGCACUUUUUGCCAACUUUGCAUAUGUGCAAAUAAUUUAGCAUCCCAGCAACAGCCAA-AACUCCA (((((........)))))..........(((.....(((((.....((.(((((.((((....(((.......)))...)))).)))))...))......))))).)))...-....... ( -15.40) >DroAna_CAF1 156 114 + 1 CCGCACCUCCCUUUCGCACAGAC-AUCCUUCUAUUUUUCGCUCUCCC-UUUGUUCGCACUUUUUGCCAACUUUGCAUAUGUGCAAAUAAUUUAGCAUCCCAACAACGACAUG-GAU---A .......(((...(((...(((.-.....))).......(((.....-.(((((.((((....(((.......)))...)))).)))))...)))..........)))...)-)).---. ( -16.20) >consensus UUGAACCUCCACUUUCAACCGACUAUCCGUUCAUUUUUGCUCCUCCCUUUUGUUCGCACUUUUUGCCAACUUUGCAUAUGUGCAAAUAAUUUAGCAUCCCAGCAAUAGCAAA_AACUUCA ...................................((((((........(((((.((((....(((.......)))...)))).)))))....((......))...))))))........ (-11.74 = -12.26 + 0.52)

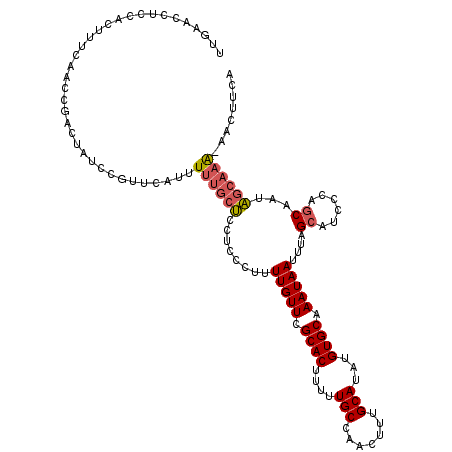
| Location | 15,753,242 – 15,753,360 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.12 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -18.26 |
| Energy contribution | -18.46 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15753242 118 + 23771897 UCCUCCCUUUUGUUCGCACUUUUUGCCAACUUUGCAUAUGUGCAAAUAAUUUAGCAUCCCAGCAAUAGCAAAAAACUUCAGCAAAAACC--AACAACAUCAGCAAGGACAAUUGUGGAUU ...((((..((((((....(((((((....((((((....)))))).......((......))....)))))))......((.......--..........))..))))))..).))).. ( -22.23) >DroSec_CAF1 4934 117 + 1 UCCUCCCUUUUGUUCGCACUUUUUGCCAACUUUGCAUAUGUGCAAAUAAUUUAGCAUCCCAGCAAUAGCAAA-AACUUCAGCAAAAACC--AACAACAUCAGCAAGGACAAUUGUGGAUU ...((((..((((((....(((((((....((((((....)))))).......((......))....)))))-)).....((.......--..........))..))))))..).))).. ( -21.93) >DroSim_CAF1 5183 117 + 1 UCCUCCCUUUUGUUCGCACUUUUUGCCAACUUUGCAUAUGUGCAAAUAAUUUAGCAUCCCAGCAAUAGCAAA-AACUUCAGCAAAAACC--AACAACAUCAGCAAGGACAAUUGUGGAUU ...((((..((((((....(((((((....((((((....)))))).......((......))....)))))-)).....((.......--..........))..))))))..).))).. ( -21.93) >DroEre_CAF1 5001 117 + 1 UCCUCCCUUUUGUUCGCACUUUUUGCCAACUUUGCAUAUGUGCAAAUAAUUUAGCAUCCCAGCAACAGCCAA-AACUCCAGCAAAAACC--AACAACAUCAGCAAGGACAAUUGUGGAUU ...((((..((((((((((....(((.......)))...))))..........((......)).........-................--..............))))))..).))).. ( -20.10) >DroAna_CAF1 195 115 + 1 CUCUCCC-UUUGUUCGCACUUUUUGCCAACUUUGCAUAUGUGCAAAUAAUUUAGCAUCCCAACAACGACAUG-GAU---AACAACAACCACACUAGCAGGAGCAAGGACAAUUGUGGAUU ...((((-.((((((((((....(((.......)))...))))..........((((((............)-)))---........((.........)).))..))))))..).))).. ( -20.80) >consensus UCCUCCCUUUUGUUCGCACUUUUUGCCAACUUUGCAUAUGUGCAAAUAAUUUAGCAUCCCAGCAAUAGCAAA_AACUUCAGCAAAAACC__AACAACAUCAGCAAGGACAAUUGUGGAUU ...((((..((((((((((....(((.......)))...))))..........((......))..........................................))))))..).))).. (-18.26 = -18.46 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:16 2006