| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,688,920 – 1,689,025 |
| Length | 105 |
| Max. P | 0.556253 |

| Location | 1,688,920 – 1,689,025 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.50 |
| Mean single sequence MFE | -21.93 |
| Consensus MFE | -12.66 |
| Energy contribution | -13.55 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.45 |
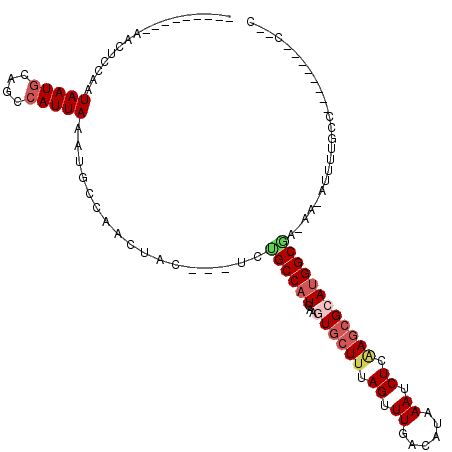
| Structure conservation index | 0.58 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
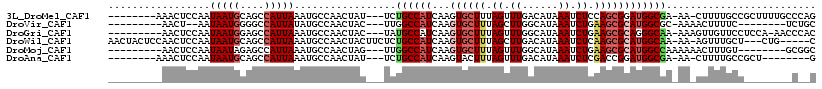
>3L_DroMel_CAF1 1688920 105 - 23771897 --------AAACUCCAAUAAUGCAGCCAUUAAAUGCCAACUAU---UCUGCCAUCAAGUGCUUUAGUUUGACAUAAAUCUCCAGCGGAUGGCGA-AA-CUUUUGCCGCUUUUGCCCAG --------.............(((((........))......(---((.((((((....(((..((.((......)).))..))).))))))))-).-....))).((....)).... ( -16.30) >DroVir_CAF1 61845 95 - 1 ---------AACU--AAUAAUGGGGCCAUUAUAUGCCAACUAC---UUGGCCAUCAAGUGCUUUAGCUUGGCAUAAAUCUGAAGCGCAUGGCGC-AAAACUUUUC--------UCUGC ---------....--.......(.(((((.....(((((....---)))))......(((((((((.((......)).)))))))))))))).)-..........--------..... ( -26.10) >DroGri_CAF1 59733 104 - 1 ---------AACUCCAAUAAUGGAGCCAUUAAAUGCCAACUAC---UAUGCCAUCAAGUGCUUUAGUUUGGCAUAAAUCUGAAGCGCAGGGCAA-AAAGUUGUUCCUCCA-AACCCAC ---------..(((((....)))))...........(((((..---..((((.....(((((((((.((......)).)))))))))..)))).-..)))))........-....... ( -26.40) >DroWil_CAF1 63553 108 - 1 AACUACUCCAACUCCAAUAAUGCAGCCAUUAAAUGCCAACUACUUCUCUGCCAUCAAGUGCUUUAGCUUGACAUAAAUCUCAAGCGCAUGGCAA-AA-AGUUUGCU---CUG-----C .....................((((.........((.((((.......((((((...((((((.((.((......)).)).)))))))))))).-..-)))).)).---)))-----) ( -19.20) >DroMoj_CAF1 58735 98 - 1 ---------AACUCCAAUAAUAGAGCCAUUAAAUGCCAACUAG---UUGGCCAUCAAGUGCUUUAGUUUGGCAUAAAUCUGAAGCGCAUGGCCAAAAAACUUUGU--------GCGGC ---------..(((........))).........(((.((.((---((((((((...(((((((((.((......)).))))))))))))))))....)))..))--------..))) ( -28.20) >DroAna_CAF1 48871 97 - 1 --------AAACUCCAAUAAUGCAGCCAUUAAAUGCCAACUAU---UCUGCCAUCAAGUACUUUAGUUUGACAUAAAUCUCGACCGGAUGGCGA-AA-CUUUUGCCGCU--------G --------..............((((......((((.(((((.---..(((......)))...))))).).)))..((((.....))))(((((-..-...))))))))--------) ( -15.40) >consensus _________AACUCCAAUAAUGCAGCCAUUAAAUGCCAACUAC___UCUGCCAUCAAGUGCUUUAGUUUGACAUAAAUCUCAAGCGCAUGGCGA_AA_AUUUUGCC________C__C .................(((((....))))).................((((((...((((((.((.((......)).)).))))))))))))......................... (-12.66 = -13.55 + 0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:55 2006