
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,752,480 – 15,752,775 |
| Length | 295 |
| Max. P | 0.994483 |

| Location | 15,752,480 – 15,752,595 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 97.97 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -25.27 |
| Energy contribution | -25.52 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15752480 115 + 23771897 CAAAAUCGCACAUUUUCCGCACUUUUCAGUUUCCACUCUUUGGCAAUUCACAAAUUUCCACACCUACGCACACAAUUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAACAG ..................((.((((.((.((.((......(((.((((....)))).)))......(((((((......))))))))).)).)).)))).)).((((....)))) ( -26.60) >DroSec_CAF1 4198 115 + 1 CGAAAUCGCACAUUUUCCGCACUUUUCAGUUUCCACUCUUUGGCAAUUCACAAAUUUCCACACCUACGCACACAAUUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAACAG .((((........)))).((.((((.((.((.((......(((.((((....)))).)))......(((((((......))))))))).)).)).)))).)).((((....)))) ( -27.40) >DroSim_CAF1 4431 115 + 1 CCAAAUCGCACAUUUUCCGCACUUUUCAGUUUCCACUCUUUGGCAAUUCACAAAUUUCCACACCUACGCACACAAUUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAACAG ..................((.((((.((.((.((......(((.((((....)))).)))......(((((((......))))))))).)).)).)))).)).((((....)))) ( -26.60) >DroEre_CAF1 4167 115 + 1 CAAAAUCGCACAUUUUCCGCACUUUUCAGUUUCCACUCUUUGCCAAUUCACAAAUUUCCACACCUACGCACACAAUUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAGCAA .......((.........((((.....(((....)))..(((((((((....))))..........(((((((......)))))))))))))))).....((......)).)).. ( -26.30) >consensus CAAAAUCGCACAUUUUCCGCACUUUUCAGUUUCCACUCUUUGGCAAUUCACAAAUUUCCACACCUACGCACACAAUUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAACAG .......(((((..(((.((((((.((.............(((.((((....)))).)))......(((((((......))))))))).))))))...)))...))).))..... (-25.27 = -25.52 + 0.25)

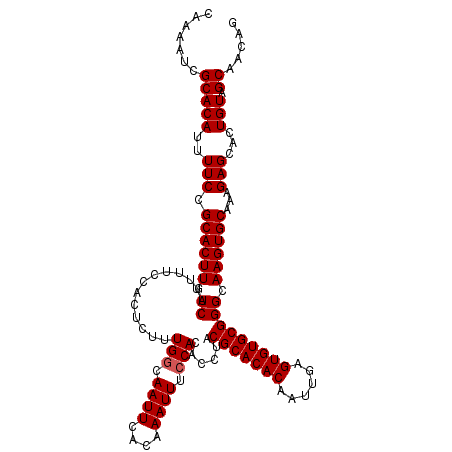
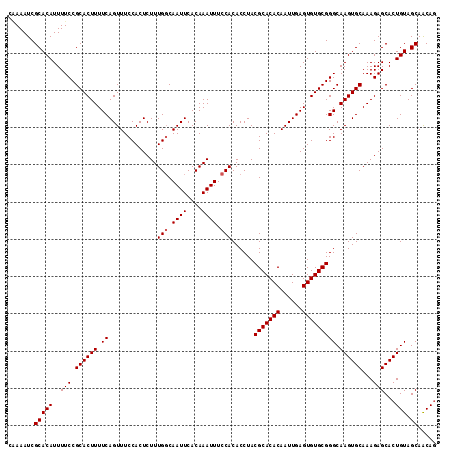
| Location | 15,752,480 – 15,752,595 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 97.97 |
| Mean single sequence MFE | -36.05 |
| Consensus MFE | -33.95 |
| Energy contribution | -34.45 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15752480 115 - 23771897 CUGUUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAAUUGUGUGCGUAGGUGUGGAAAUUUGUGAAUUGCCAAAGAGUGGAAACUGAAAAGUGCGGAAAAUGUGCGAUUUUG ..(((((.(((..(((((((((((((((..((((((......))))))))))))))((.((((....)))).)))))))))....(((....)))........)))))))).... ( -36.10) >DroSec_CAF1 4198 115 - 1 CUGUUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAAUUGUGUGCGUAGGUGUGGAAAUUUGUGAAUUGCCAAAGAGUGGAAACUGAAAAGUGCGGAAAAUGUGCGAUUUCG ((((....)))).(((((((((((((((..((((((......))))))))))))))((.((((....)))).)))))))))(....).((((.(..((.....))..)..)))). ( -37.00) >DroSim_CAF1 4431 115 - 1 CUGUUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAAUUGUGUGCGUAGGUGUGGAAAUUUGUGAAUUGCCAAAGAGUGGAAACUGAAAAGUGCGGAAAAUGUGCGAUUUGG ..(((((.(((..(((((((((((((((..((((((......))))))))))))))((.((((....)))).)))))))))....(((....)))........)))))))).... ( -36.10) >DroEre_CAF1 4167 115 - 1 UUGCUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAAUUGUGUGCGUAGGUGUGGAAAUUUGUGAAUUGGCAAAGAGUGGAAACUGAAAAGUGCGGAAAAUGUGCGAUUUUG ...((((.((...((((((..(((((((..((((((......)))))))))))))..))..(((((......)))))))))(....)......))))))................ ( -35.00) >consensus CUGUUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAAUUGUGUGCGUAGGUGUGGAAAUUUGUGAAUUGCCAAAGAGUGGAAACUGAAAAGUGCGGAAAAUGUGCGAUUUUG ..(((((.(((..(((((((((((((((..((((((......))))))))))))))((.((((....)))).)))))))))....(((....)))........)))))))).... (-33.95 = -34.45 + 0.50)

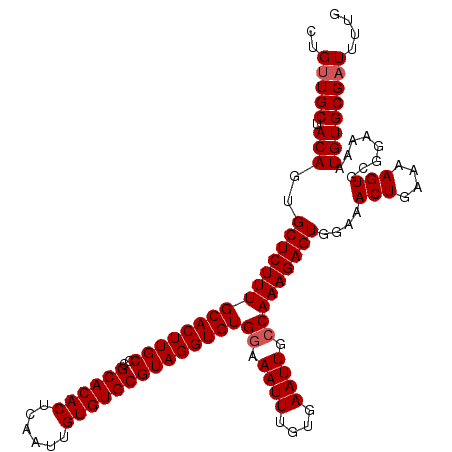
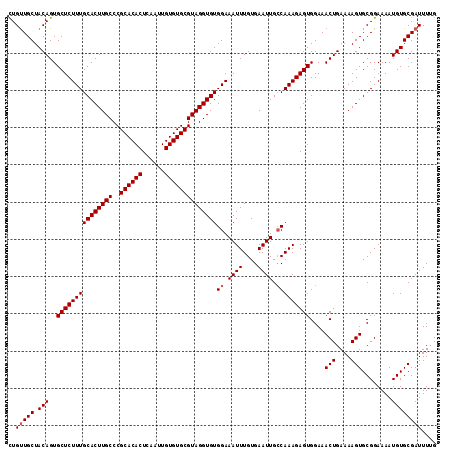
| Location | 15,752,520 – 15,752,632 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 95.29 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -29.48 |
| Energy contribution | -29.73 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15752520 112 + 23771897 UGGCAAUUCACAAAUUUCCACACCUACGCACACAAUUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAACAGCAACAA---CAACAAAGAAAUUGUUAAUGCCAUUUGACAG (((((.((.((((.((((........(((((((......))))))).((.(((((.....)))))...))...........---.......)))))))).)))))))........ ( -31.40) >DroSec_CAF1 4238 109 + 1 UGGCAAUUCACAAAUUUCCACACCUACGCACACAAUUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAACAGCAAC------AACAAAGAAAUUGUUAAUGCCAUUUGACAG (((((.((.((((.((((........(((((((......))))))).((.(((((.....)))))...)).........------......)))))))).)))))))........ ( -31.40) >DroSim_CAF1 4471 115 + 1 UGGCAAUUCACAAAUUUCCACACCUACGCACACAAUUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAACAGCAACAACAACAACAAAGAAAUUGUUAAUGCCAUUUGACAG (((((.((.((((.((((........(((((((......))))))).((.(((((.....)))))...)).....................)))))))).)))))))........ ( -31.40) >DroEre_CAF1 4207 109 + 1 UGCCAAUUCACAAAUUUCCACACCUACGCACACAAUUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAGCAACAA------CAACAAAGAAAUUGUUAAUGCCAUUUGACAG ..........(((((...........(((((((......)))))))(((((((((.....))))).............------.(((((.....)))))..))))))))).... ( -28.20) >consensus UGGCAAUUCACAAAUUUCCACACCUACGCACACAAUUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAACAGCAAC_____CAACAAAGAAAUUGUUAAUGCCAUUUGACAG (((((.((.((((.((((........(((((((......))))))).((.(((((.....)))))...)).....................)))))))).)))))))........ (-29.48 = -29.73 + 0.25)

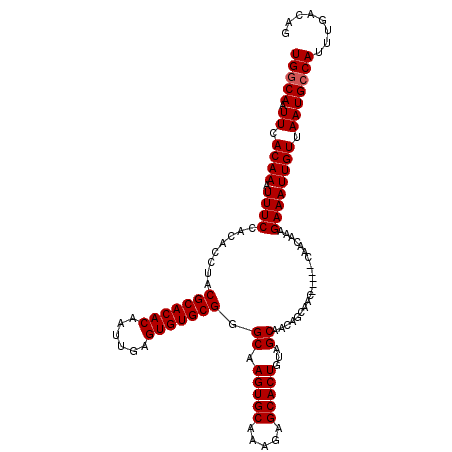
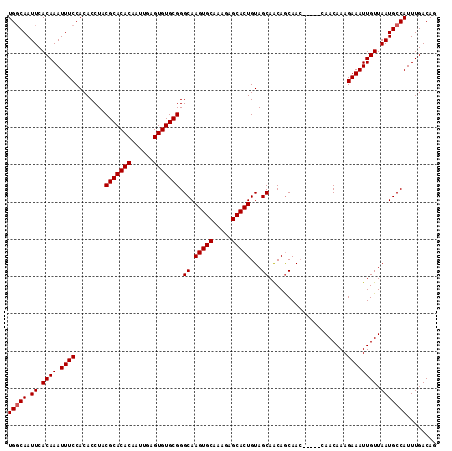
| Location | 15,752,520 – 15,752,632 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 95.29 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -32.04 |
| Energy contribution | -32.35 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.63 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15752520 112 - 23771897 CUGUCAAAUGGCAUUAACAAUUUCUUUGUUG---UUGUUGCUGUUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAAUUGUGUGCGUAGGUGUGGAAAUUUGUGAAUUGCCA ((((..(((((((.(((((((......))))---))).)))))))...)))).....((..(((((((..((((((......)))))))))))))..))................ ( -38.10) >DroSec_CAF1 4238 109 - 1 CUGUCAAAUGGCAUUAACAAUUUCUUUGUU------GUUGCUGUUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAAUUGUGUGCGUAGGUGUGGAAAUUUGUGAAUUGCCA ((((..(((((((.((((((.....)))))------).)))))))...)))).....((..(((((((..((((((......)))))))))))))..))................ ( -36.80) >DroSim_CAF1 4471 115 - 1 CUGUCAAAUGGCAUUAACAAUUUCUUUGUUGUUGUUGUUGCUGUUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAAUUGUGUGCGUAGGUGUGGAAAUUUGUGAAUUGCCA ((((..(((((((.(((((((.........))))))).)))))))...)))).....((..(((((((..((((((......)))))))))))))..))................ ( -37.10) >DroEre_CAF1 4207 109 - 1 CUGUCAAAUGGCAUUAACAAUUUCUUUGUUG------UUGUUGCUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAAUUGUGUGCGUAGGUGUGGAAAUUUGUGAAUUGGCA ..(((((..((((.(((((((......))))------))).)))).(((((((((.....))))).((((((((((......)))))))..)))))))...........))))). ( -39.10) >consensus CUGUCAAAUGGCAUUAACAAUUUCUUUGUUG_____GUUGCUGUUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAAUUGUGUGCGUAGGUGUGGAAAUUUGUGAAUUGCCA ........(((((...(((((((((..............(((((....))))).......((((((((..((((((......))))))))))))))))))).))))....))))) (-32.04 = -32.35 + 0.31)

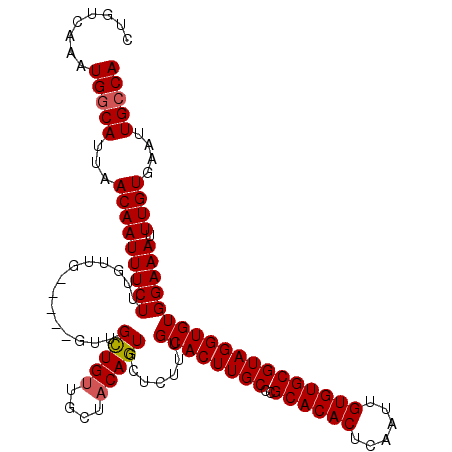
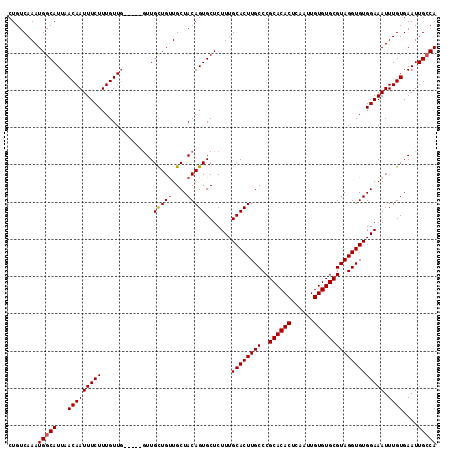
| Location | 15,752,555 – 15,752,672 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.91 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -29.54 |
| Energy contribution | -29.35 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15752555 117 + 23771897 UUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAACAGCAACAA---CAACAAAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAG ((((((((...(((((((((.....)))))((.((....)).))..---.(((((.....)))))..))))...(((((((((.(.......))))))))))..))))))))........ ( -30.70) >DroSec_CAF1 4273 114 + 1 UUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAACAGCAAC------AACAAAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAG ((((((((...(((((((((.....)))))((.((....)).))------(((((.....)))))..))))...(((((((((.(.......))))))))))..))))))))........ ( -30.70) >DroSim_CAF1 4506 120 + 1 UUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAACAGCAACAACAACAACAAAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAG ((((((((...(((((((((.....)))))((.((....)).))......(((((.....)))))..))))...(((((((((.(.......))))))))))..))))))))........ ( -30.70) >DroEre_CAF1 4242 114 + 1 UUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAGCAACAA------CAACAAAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAG ....((.(((..((.(((((.....)))))...)).)))...)------)...........(((((((((....(((((((((.(.......))))))))))..))).))))))...... ( -30.50) >consensus UUGAGUGUGCGGGCAAGUGCAAAGAGCACUGUAGCAACAGCAAC_____CAACAAAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAG ....((.(((..(((.((((.....))))))).)))...))....................(((((((((....(((((((((.(.......))))))))))..))).))))))...... (-29.54 = -29.35 + -0.19)

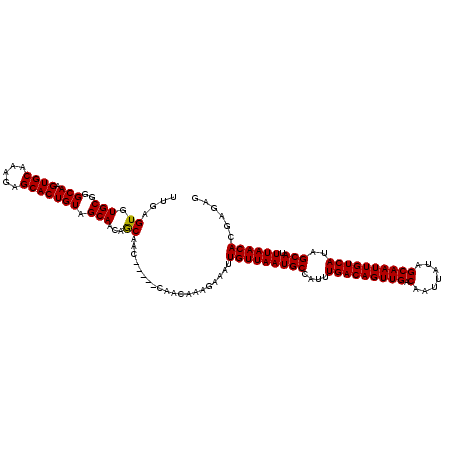
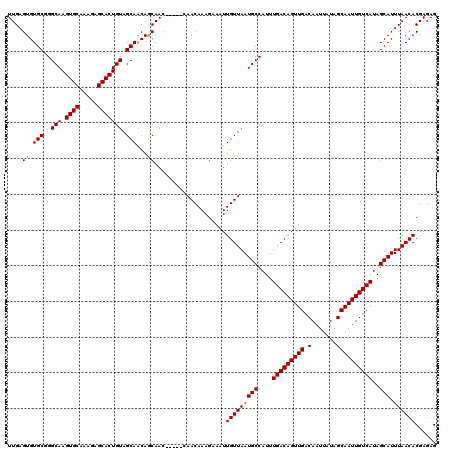
| Location | 15,752,555 – 15,752,672 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.91 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -26.61 |
| Energy contribution | -26.43 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15752555 117 - 23771897 CUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUUUGUUG---UUGUUGCUGUUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAA .....(((((((.(((..(((((((((...)))))))))(((((..(((((((.(((((((......))))---))).)))))))...))))).......))).)))...))))...... ( -32.30) >DroSec_CAF1 4273 114 - 1 CUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUUUGUU------GUUGCUGUUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAA .....(((((((.(((..(((((((((...)))))))))(((((..(((((((.((((((.....)))))------).)))))))...))))).......))).)))...))))...... ( -31.00) >DroSim_CAF1 4506 120 - 1 CUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUUUGUUGUUGUUGUUGCUGUUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAA .....(((((((.(((..(((((((((...)))))))))(((((..(((((((.(((((((.........))))))).)))))))...))))).......))).)))...))))...... ( -31.30) >DroEre_CAF1 4242 114 - 1 CUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUUUGUUG------UUGUUGCUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAA .....((((..((((((((((((.((((.......).))).)))))..)))))))((((((......))))------))...((.((...(((((.....))))).))..)))))).... ( -29.10) >consensus CUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUUUGUUG_____GUUGCUGUUGCUACAGUGCUCUUUGCACUUGCCCGCACACUCAA .....((((..((((((((((((.((((.......).))).)))))..)))))))(((((.....))))).........((....((...(((((.....))))).))..)))))).... (-26.61 = -26.43 + -0.19)

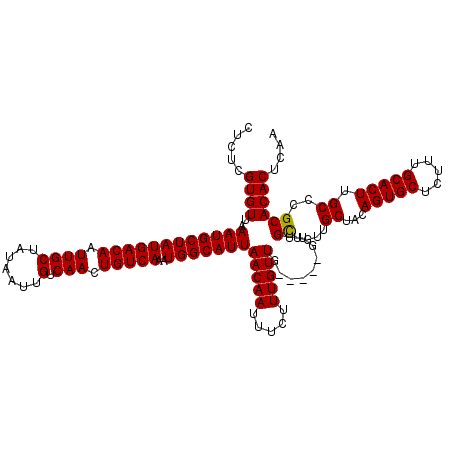
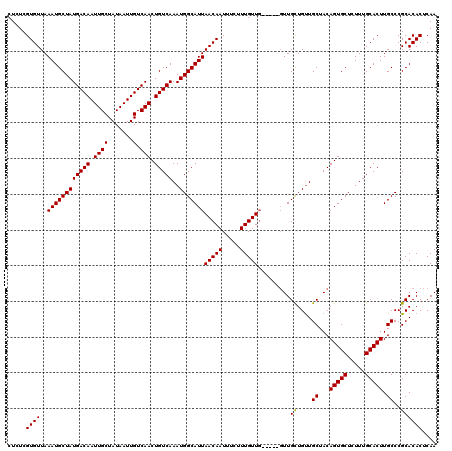
| Location | 15,752,595 – 15,752,712 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.56 |
| Mean single sequence MFE | -23.02 |
| Consensus MFE | -19.65 |
| Energy contribution | -20.65 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15752595 117 + 23771897 CAACAA---CAACAAAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGCUGAGCACUAUACGAAAUGCGAAAACGGAUGGAUGAAAGA ......---.(((((.....)))))....(((((((.....((((((((.....))))))))..((((((......((.((...)).)).....)))))).....)))))))........ ( -23.40) >DroSec_CAF1 4313 104 + 1 CAAC------AACAAAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGA----------GAGAAAUGCGGAAGGGGAUGGAUGAAAGA ....------(((((.....)))))....((((((((((((((.(.......)))))))))...((((((..(.(....).----------)..))))))......))))))........ ( -22.90) >DroSim_CAF1 4546 120 + 1 CAACAACAACAACAAAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGCUGAGCACUAUACGAAAUGCGGAAAGGGAUGGAUGAAAGA ..........(((((.....)))))....((((((((((((((.(.......)))))))))...((((((......((.((...)).)).....))))))......))))))........ ( -22.90) >DroEre_CAF1 4282 114 + 1 CAA------CAACAAAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGCUGAGCACUAUACGAAAUGCGGAAAGGGAUGGAUGAAAUA ...------.(((((.....)))))....((((((((((((((.(.......)))))))))...((((((......((.((...)).)).....))))))......))))))........ ( -22.90) >consensus CAAC_____CAACAAAGAAAUUGUUAAUGCCAUUUGACAGUUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGCUGAGCACUAUACGAAAUGCGGAAAGGGAUGGAUGAAAGA ..........(((((.....)))))....((((((((((((((.(.......)))))))))...((((((......((.((...)).)).....))))))......))))))........ (-19.65 = -20.65 + 1.00)


| Location | 15,752,595 – 15,752,712 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.56 |
| Mean single sequence MFE | -22.53 |
| Consensus MFE | -16.38 |
| Energy contribution | -17.62 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15752595 117 - 23771897 UCUUUCAUCCAUCCGUUUUCGCAUUUCGUAUAGUGCUCAGCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUUUGUUG---UUGUUG ............((((((..((((((..(((((.((...)).)).)))..))))))..(((((((((...)))))))))......))))))((.(((((((......))))---))).)) ( -23.90) >DroSec_CAF1 4313 104 - 1 UCUUUCAUCCAUCCCCUUCCGCAUUUCUC----------UCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUUUGUU------GUUG ....................((((((..(----------........)..))))))..(((((((((...)))))))))..((((....)))).(((((((......)))------)))) ( -18.70) >DroSim_CAF1 4546 120 - 1 UCUUUCAUCCAUCCCUUUCCGCAUUUCGUAUAGUGCUCAGCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUUUGUUGUUGUUGUUG ....................((((((..(((((.((...)).)).)))..))))))..(((((((((...))))))))).......((..(((.((((((.....)))))).)))..)). ( -25.10) >DroEre_CAF1 4282 114 - 1 UAUUUCAUCCAUCCCUUUCCGCAUUUCGUAUAGUGCUCAGCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUUUGUUG------UUG ....................(((((......))))).((((.....((((((.((((((((((.((((.......).))).)))))..)))))))))))........))))------... ( -22.42) >consensus UCUUUCAUCCAUCCCUUUCCGCAUUUCGUAUAGUGCUCAGCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAACUGUCAAAUGGCAUUAACAAUUUCUUUGUUG_____GUUG ....................(((((......)))))..........((((((.((((((((((.((((.......).))).)))))..)))))))))))..................... (-16.38 = -17.62 + 1.25)

| Location | 15,752,632 – 15,752,752 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15752632 120 + 23771897 UUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGCUGAGCACUAUACGAAAUGCGAAAACGGAUGGAUGAAAGAGUCCAUUGUUGUCUGCACAGAGAAAAAAGAAAUGGCAAAU .((((((((.....))))))))..((((((......((.((...)).)).....))))))((.((((.((((((......)))))))))).))(((.((.............)))))... ( -25.82) >DroSec_CAF1 4347 110 + 1 UUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGA----------GAGAAAUGCGGAAGGGGAUGGAUGAAAGAGUCCAUUGUUGUCUGCACGGAAAAAAAAGAAAUGGCAAAU .((((((((.....))))))))...(((((....(....).----------......((((((..(.(((((((......))))))).)..))))))............)))))...... ( -28.00) >DroSim_CAF1 4586 120 + 1 UUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGCUGAGCACUAUACGAAAUGCGGAAAGGGAUGGAUGAAAGAGUCCAUUGUUGUGUGCACGGAGAAAAAAGAAAUGGCAAAU .((((((((.....))))))))...(((((..........(((.((((....((.....))...((.(((((((......))))))).))..)))).))).........)))))...... ( -26.81) >DroEre_CAF1 4316 119 + 1 UUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGCUGAGCACUAUACGAAAUGCGGAAAGGGAUGGAUGAAAUAGUCCAUUGUUGUCUGCACGUAGAAA-AACAAAUGCCAAAA .((((((((.....))))))))..((((((..(((....).)).......((((...((((((.((.(((((((......))))))).)).))))))))))....-...))))))..... ( -33.20) >consensus UUGACAAUUAUAGCAAUUGUCAUAGCAUUUAACACGAGAGCUGAGCACUAUACGAAAUGCGGAAAGGGAUGGAUGAAAGAGUCCAUUGUUGUCUGCACGGAGAAAAAAGAAAUGGCAAAU .((((((((.....))))))))..((((((....(....).................((((((.((.(((((((......))))))).)).))))))............))))).).... (-23.34 = -23.90 + 0.56)

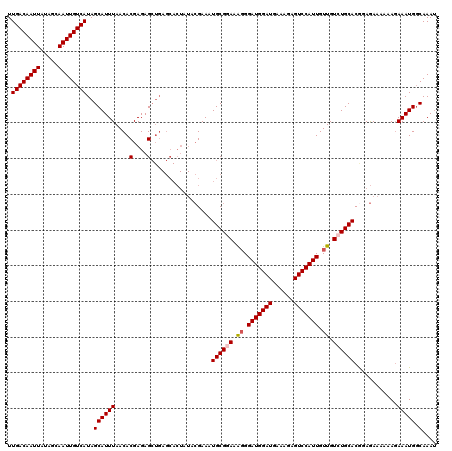
| Location | 15,752,632 – 15,752,752 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -21.97 |
| Consensus MFE | -16.20 |
| Energy contribution | -17.45 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15752632 120 - 23771897 AUUUGCCAUUUCUUUUUUCUCUGUGCAGACAACAAUGGACUCUUUCAUCCAUCCGUUUUCGCAUUUCGUAUAGUGCUCAGCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAA ...(((((.............)).)))((.(((.(((((........)))))..))).))((((((..(((((.((...)).)).)))..))))))..(((((((((...))))))))). ( -23.42) >DroSec_CAF1 4347 110 - 1 AUUUGCCAUUUCUUUUUUUUCCGUGCAGACAACAAUGGACUCUUUCAUCCAUCCCCUUCCGCAUUUCUC----------UCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAA .((((((...............).))))).....(((((........)))))........((((((..(----------........)..))))))..(((((((((...))))))))). ( -17.66) >DroSim_CAF1 4586 120 - 1 AUUUGCCAUUUCUUUUUUCUCCGUGCACACAACAAUGGACUCUUUCAUCCAUCCCUUUCCGCAUUUCGUAUAGUGCUCAGCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAA .....................(((..((((....(((((........)))))........(((((......))))).........))))...)))...(((((((((...))))))))). ( -19.60) >DroEre_CAF1 4316 119 - 1 UUUUGGCAUUUGUU-UUUCUACGUGCAGACAACAAUGGACUAUUUCAUCCAUCCCUUUCCGCAUUUCGUAUAGUGCUCAGCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAA ...(((((((((..-....((((.((........(((((........)))))........(((((......)))))...))...)))).)))))))))(((((((((...))))))))). ( -27.20) >consensus AUUUGCCAUUUCUUUUUUCUCCGUGCAGACAACAAUGGACUCUUUCAUCCAUCCCUUUCCGCAUUUCGUAUAGUGCUCAGCUCUCGUGUUAAAUGCUAUGACAAUUGCUAUAAUUGUCAA ........................(((...(((((((((........)))))........(((((......)))))..........))))...)))..(((((((((...))))))))). (-16.20 = -17.45 + 1.25)

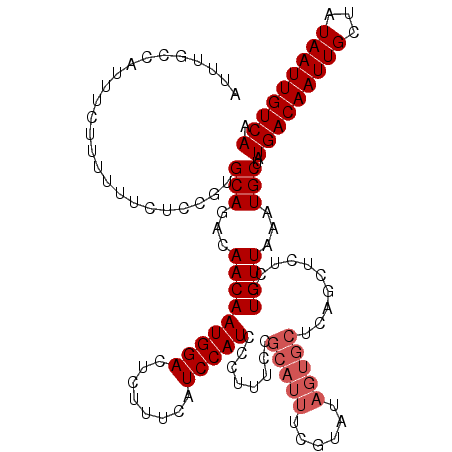
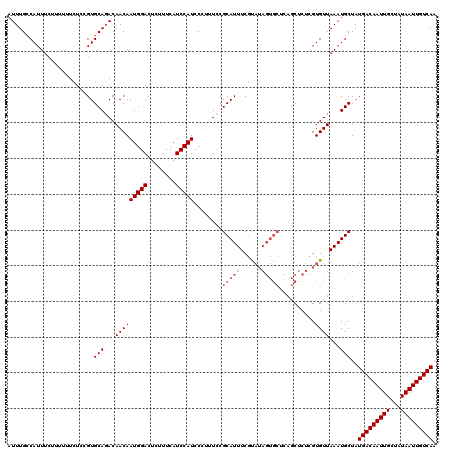
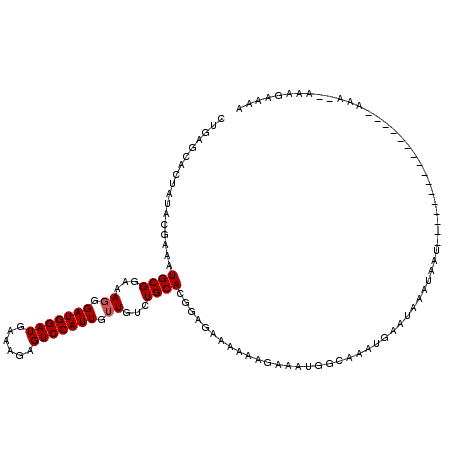
| Location | 15,752,672 – 15,752,775 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.49 |
| Mean single sequence MFE | -18.82 |
| Consensus MFE | -11.67 |
| Energy contribution | -12.00 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15752672 103 + 23771897 CUGAGCACUAUACGAAAUGCGAAAACGGAUGGAUGAAAGAGUCCAUUGUUGUCUGCACAGAGAAAAAAGAAAUGGCAAAUGAAUAAAUAAA----------------AGU-UAAGGAAAA ((.(((..............((.((((.((((((......)))))))))).))(((.((.............)))))..............----------------.))-).))..... ( -13.82) >DroSim_CAF1 4626 103 + 1 CUGAGCACUAUACGAAAUGCGGAAAGGGAUGGAUGAAAGAGUCCAUUGUUGUGUGCACGGAGAAAAAAGAAAUGGCAAAUGAAUAAAUUAU----------------ACGG-AAAGAAAA (((.((((....((.....))...((.(((((((......))))))).))..)))).)))...............................----------------....-........ ( -16.90) >DroEre_CAF1 4356 119 + 1 CUGAGCACUAUACGAAAUGCGGAAAGGGAUGGAUGAAAUAGUCCAUUGUUGUCUGCACGUAGAAA-AACAAAUGCCAAAAUAAUAAAUCAUGUUUUUCCAAAAAUCAAAACGAAUGAAAA .(((......((((...((((((.((.(((((((......))))))).)).))))))))))((((-((((....................))))))))......)))............. ( -25.75) >consensus CUGAGCACUAUACGAAAUGCGGAAAGGGAUGGAUGAAAGAGUCCAUUGUUGUCUGCACGGAGAAAAAAGAAAUGGCAAAUGAAUAAAUAAU________________AAA__AAAGAAAA .................((((...((.(((((((......))))))).))...))))............................................................... (-11.67 = -12.00 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:01:13 2006