
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
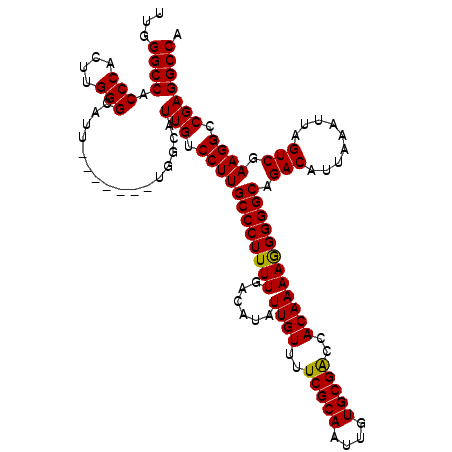
| Location | 15,750,571 – 15,750,724 |
| Length | 153 |
| Max. P | 0.974105 |

| Location | 15,750,571 – 15,750,684 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -37.60 |
| Energy contribution | -37.23 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
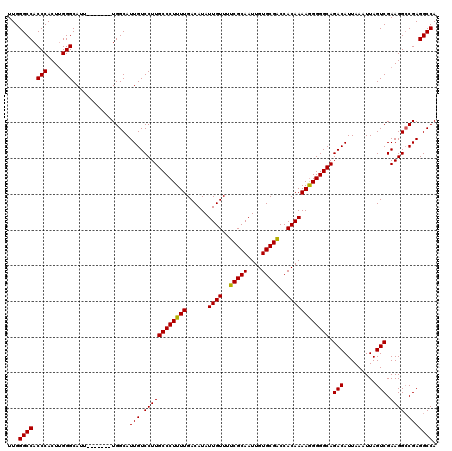
>3L_DroMel_CAF1 15750571 113 + 23771897 UUGGGCCACCCACUUGGGCAUU-------UGGCAUUGUCCUUGCCCUUUUGACAUAUUGUUUUCGCAAUUGUGCGACCACAAAAGGGGGCAGACAUUAAAUUAGUCGAAGGCCGAGGCCA ...((((.(((....)))...(-------((((....((..(((((((((......((((..(((((....)))))..)))))))))))))(((.........)))))..))))))))). ( -40.60) >DroSec_CAF1 2269 113 + 1 UUGGGCCACCCACUUGGGCAUU-------UGGCAUUGUCCUUGCCCUUUUGACAUAUUGUUUUCGCAAUUGUGCGACCACAAAAGGGGGCAGACAUUAAAUUAGUCGAAGGCCGAGGCCA ...((((.(((....)))...(-------((((....((..(((((((((......((((..(((((....)))))..)))))))))))))(((.........)))))..))))))))). ( -40.60) >DroSim_CAF1 2500 113 + 1 UUGGGCCACCCACUCGGGCAUU-------UGGCAUUGUCCUUGCCCUUUUGACAUAUUGUUUUCGCAAUUGUGCGACCACAAAAGGGGGCAGACAUUAAAUUAGUCGAAGGCCGAGGCCA ...((((.(((....)))...(-------((((....((..(((((((((......((((..(((((....)))))..)))))))))))))(((.........)))))..))))))))). ( -40.30) >DroEre_CAF1 2252 119 + 1 UUGGGCCACCCACUUGGGCAUUUGGCAUUUGGCAUUGUCCUUGCCCUUUUGACAUAUUGUUUUCGCAAUUGUGCGGCCACAAAAAGGGGCAGACAUUAAAUUAGUCGAAGG-CGAGGCCA ....(((((((....)))....))))...((((.(((.((((((((((((......((((..(((((....)))))..)))))))))))).(((.........))).))))-))).)))) ( -40.40) >consensus UUGGGCCACCCACUUGGGCAUU_______UGGCAUUGUCCUUGCCCUUUUGACAUAUUGUUUUCGCAAUUGUGCGACCACAAAAGGGGGCAGACAUUAAAUUAGUCGAAGGCCGAGGCCA ...((((.(((....)))................(((.((((((((((((......((((..(((((....)))))..)))))))))))).(((.........))).)))).))))))). (-37.60 = -37.23 + -0.37)

| Location | 15,750,571 – 15,750,684 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -33.46 |
| Energy contribution | -33.52 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15750571 113 - 23771897 UGGCCUCGGCCUUCGACUAAUUUAAUGUCUGCCCCCUUUUGUGGUCGCACAAUUGCGAAAACAAUAUGUCAAAAGGGCAAGGACAAUGCCA-------AAUGCCCAAGUGGGUGGCCCAA .(((....)))...(((.........))).((((((..((((..(((((....)))))..))))..........(((((.((......)).-------..)))))....))).))).... ( -36.80) >DroSec_CAF1 2269 113 - 1 UGGCCUCGGCCUUCGACUAAUUUAAUGUCUGCCCCCUUUUGUGGUCGCACAAUUGCGAAAACAAUAUGUCAAAAGGGCAAGGACAAUGCCA-------AAUGCCCAAGUGGGUGGCCCAA .(((....)))...(((.........))).((((((..((((..(((((....)))))..))))..........(((((.((......)).-------..)))))....))).))).... ( -36.80) >DroSim_CAF1 2500 113 - 1 UGGCCUCGGCCUUCGACUAAUUUAAUGUCUGCCCCCUUUUGUGGUCGCACAAUUGCGAAAACAAUAUGUCAAAAGGGCAAGGACAAUGCCA-------AAUGCCCGAGUGGGUGGCCCAA .(((....)))...(((.........))).((((((..((((..(((((....)))))..))))..........(((((.((......)).-------..)))))....))).))).... ( -36.80) >DroEre_CAF1 2252 119 - 1 UGGCCUCG-CCUUCGACUAAUUUAAUGUCUGCCCCUUUUUGUGGCCGCACAAUUGCGAAAACAAUAUGUCAAAAGGGCAAGGACAAUGCCAAAUGCCAAAUGCCCAAGUGGGUGGCCCAA .((((...-.....(((.........))).(((((((.((((...((((....))))...))))..........(((((.((.((........))))...)))))))).))))))))... ( -33.10) >consensus UGGCCUCGGCCUUCGACUAAUUUAAUGUCUGCCCCCUUUUGUGGUCGCACAAUUGCGAAAACAAUAUGUCAAAAGGGCAAGGACAAUGCCA_______AAUGCCCAAGUGGGUGGCCCAA .((((....................(((((...((((((((((.(((((....)))))...))......))))))))...)))))................((((....))))))))... (-33.46 = -33.52 + 0.06)



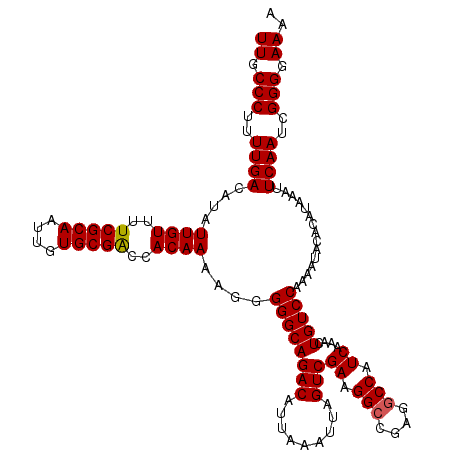
| Location | 15,750,604 – 15,750,724 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -32.41 |
| Consensus MFE | -30.99 |
| Energy contribution | -31.05 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15750604 120 + 23771897 UUGCCCUUUUGACAUAUUGUUUUCGCAAUUGUGCGACCACAAAAGGGGGCAGACAUUAAAUUAGUCGAAGGCCGAGGCCAUCAAACUGUCCAAAAUACACAUAAAUUCAAUCGGGGAAAA ((.(((..((((....((((..(((((....)))))..))))....((((((((.........)))((.(((....))).))....)))))...............))))..))).)).. ( -32.90) >DroSec_CAF1 2302 120 + 1 UUGCCCUUUUGACAUAUUGUUUUCGCAAUUGUGCGACCACAAAAGGGGGCAGACAUUAAAUUAGUCGAAGGCCGAGGCCAUCAAACUGUCCAAAAUACACAUAAAUUCAAUCGGGGAAAA ((.(((..((((....((((..(((((....)))))..))))....((((((((.........)))((.(((....))).))....)))))...............))))..))).)).. ( -32.90) >DroSim_CAF1 2533 120 + 1 UUGCCCUUUUGACAUAUUGUUUUCGCAAUUGUGCGACCACAAAAGGGGGCAGACAUUAAAUUAGUCGAAGGCCGAGGCCAUCAAACUGUCCAAAAUACACAUAAAUUCAAUCGGGGAAAA ((.(((..((((....((((..(((((....)))))..))))....((((((((.........)))((.(((....))).))....)))))...............))))..))).)).. ( -32.90) >DroEre_CAF1 2292 119 + 1 UUGCCCUUUUGACAUAUUGUUUUCGCAAUUGUGCGGCCACAAAAAGGGGCAGACAUUAAAUUAGUCGAAGG-CGAGGCCAUCGAGCUGUCCAAAAUACACAUAAAUUCAAUCGGGGAAUA ((.(((..((((....((((..(((((....)))))..))))....((((((............((((.((-(...))).)))).))))))...............))))..))).)).. ( -30.92) >consensus UUGCCCUUUUGACAUAUUGUUUUCGCAAUUGUGCGACCACAAAAGGGGGCAGACAUUAAAUUAGUCGAAGGCCGAGGCCAUCAAACUGUCCAAAAUACACAUAAAUUCAAUCGGGGAAAA ((.(((..((((....((((..(((((....)))))..))))....((((((((.........)))((.(((....))).))....)))))...............))))..))).)).. (-30.99 = -31.05 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:59 2006