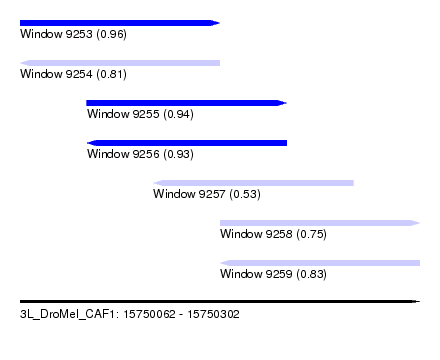
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,750,062 – 15,750,302 |
| Length | 240 |
| Max. P | 0.955216 |

| Location | 15,750,062 – 15,750,182 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -43.48 |
| Consensus MFE | -33.05 |
| Energy contribution | -33.42 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15750062 120 + 23771897 UUGGGCUUAACCUCGCCUGACCUACUCCAGAACGGAGCCGAAGCCGUUGGAAAAGCAGGAGCAGAAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUU (..(((........)))..).............((((.....((((((.....))).(((((((..(((....)))..))))))).))).((((((((......)))))))))))).... ( -41.70) >DroSec_CAF1 1766 120 + 1 UUGGACUUAACCUCGCCUGGCCUACUCCAGAAUGGAGCCGGAGCAGUUGGAAAAGCUGGAGCAGAAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUU .((((.....((......)).....))))((((((((((((((((((((.....((.(((.......)))))...))))))))))))...((((((((......)))))))))))))))) ( -44.90) >DroSim_CAF1 1997 120 + 1 UUGGGCUUAACCUCGCCUGACCUACUCCAGAAUGGAGCCGGAGCAGUUGGAAAAGCUAUAGCAGAAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUU (..(((........)))..).........((((((((((((((((((((((...((....)).....))))......))))))))))...((((((((......)))))))))))))))) ( -44.90) >DroEre_CAF1 1758 102 + 1 UCGGGCUUAACCUCGCCUGACCUACUCCAGAAUGGGGCAGG------------------AGCAGCAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUU ((((((........)))))).........(((((((((.((------------------((((((.(((....))).))))))))..)).((((((((......)))))))).))))))) ( -42.40) >consensus UUGGGCUUAACCUCGCCUGACCUACUCCAGAAUGGAGCCGGAGCAGUUGGAAAAGCUGGAGCAGAAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUU ..(((.........(((((.((..((((.....))))..)).................((((((..(((....)))..))))))))))).((((((((......)))))))).))).... (-33.05 = -33.42 + 0.38)


| Location | 15,750,062 – 15,750,182 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -41.06 |
| Consensus MFE | -31.17 |
| Energy contribution | -31.67 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15750062 120 - 23771897 AAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUUCUGCUCCUGCUUUUCCAACGGCUUCGGCUCCGUUCUGGAGUAGGUCAGGCGAGGUUAAGCCCAA ...(((((((((((((......)))))))..((..(((((((...((....))...))))))).)).))))))..(((((.(((..(((.((((......)))))))..))))))))... ( -41.80) >DroSec_CAF1 1766 120 - 1 AAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUUCUGCUCCAGCUUUUCCAACUGCUCCGGCUCCAUUCUGGAGUAGGCCAGGCGAGGUUAAGUCCAA .(((((((.(((((((......)))))))....(((((((((......((((((....).))).))........)))))))))))))))).((((.(..(((......)))..).)))). ( -45.24) >DroSim_CAF1 1997 120 - 1 AAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUUCUGCUAUAGCUUUUCCAACUGCUCCGGCUCCAUUCUGGAGUAGGUCAGGCGAGGUUAAGCCCAA ...(((((((.(((((......)))))))))(((.((((.(((.((..((((....)))).)).))).))))..(((((((((.......)))))))))....)))..........))). ( -41.80) >DroEre_CAF1 1758 102 - 1 AAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUGCUGCU------------------CCUGCCCCAUUCUGGAGUAGGUCAGGCGAGGUUAAGCCCGA ....(.((((.(((((......))))))))).)((((((((((..((....))..))))))------------------))(((.((.....)).)))))((.(((........))).)) ( -35.40) >consensus AAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUUCUGCUCCAGCUUUUCCAACUGCUCCGGCUCCAUUCUGGAGUAGGUCAGGCGAGGUUAAGCCCAA ...(((((((.(((((......)))))))))(((.(((((((...((....))...)))))))................((.(((((.....))))).))...)))..........))). (-31.17 = -31.67 + 0.50)



| Location | 15,750,102 – 15,750,222 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -28.40 |
| Energy contribution | -31.65 |
| Covariance contribution | 3.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15750102 120 + 23771897 AAGCCGUUGGAAAAGCAGGAGCAGAAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUUUUCAUAUGCGCAUUAAGCGCCUUUUCCACAUUUCAAUUUG .....((.(((((((..(((((((..(((....)))..))))))).((..((((((((......))))))))..))...........((((.....)))))))))))))........... ( -41.60) >DroSec_CAF1 1806 120 + 1 GAGCAGUUGGAAAAGCUGGAGCAGAAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUUUUCAUAUGCGCAUUAAGCGCCUUUUCCACAUUUCAAUUUG (((..((.(((((((.((((((((..(((....)))..))))))))((..((((((((......))))))))..))...........((((.....)))))))))))))..)))...... ( -41.90) >DroSim_CAF1 2037 120 + 1 GAGCAGUUGGAAAAGCUAUAGCAGAAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUUUUCAUAUGCGCAUUAAGCGCCUUUUCCACAUUUCAAUUUG (((..((.(((((((....(((((..(((....)))..)))))...((..((((((((......))))))))..))...........((((.....)))))))))))))..)))...... ( -34.20) >DroEre_CAF1 1798 102 + 1 G------------------AGCAGCAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUUUUCAUAUGCACAUUAAGCGCCUUUUCCACAUUUCAAUUUG (------------------((((((.(((....))).))))))).((((.((((((((......))))))))...............((.......)))))).................. ( -27.80) >consensus GAGCAGUUGGAAAAGCUGGAGCAGAAGUCCGCAGAUGGCUGCUCCGGGCAGAAGGAAAAUGCAAUUUCCUUUCUCCAUUUUUCAUAUGCGCAUUAAGCGCCUUUUCCACAUUUCAAUUUG .......((((((((..(((((((..(((....)))..))))))).((..((((((((......))))))))..))...........((((.....))))))))))))............ (-28.40 = -31.65 + 3.25)

| Location | 15,750,102 – 15,750,222 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -29.01 |
| Energy contribution | -31.45 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15750102 120 - 23771897 CAAAUUGAAAUGUGGAAAAGGCGCUUAAUGCGCAUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUUCUGCUCCUGCUUUUCCAACGGCUU ............((((((((((((.....))))...........((((((.(((((......))))))))).)).(((((((...((....))...)))))))..))))))))....... ( -39.30) >DroSec_CAF1 1806 120 - 1 CAAAUUGAAAUGUGGAAAAGGCGCUUAAUGCGCAUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUUCUGCUCCAGCUUUUCCAACUGCUC ............((((((((((((.....))))...........((((((.(((((......))))))))).)).(((((((...((....))...)))))))..))))))))....... ( -39.80) >DroSim_CAF1 2037 120 - 1 CAAAUUGAAAUGUGGAAAAGGCGCUUAAUGCGCAUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUUCUGCUAUAGCUUUUCCAACUGCUC ............((((((((((((.....)))).((((....((((...(((((((......)))))))((((.....))))))))..((((....)))))))).))))))))....... ( -34.40) >DroEre_CAF1 1798 102 - 1 CAAAUUGAAAUGUGGAAAAGGCGCUUAAUGUGCAUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUGCUGCU------------------C ............(((.....((((.....))))...........(.((((.(((((......))))))))).))))(((((((..((....))..))))))------------------) ( -28.40) >consensus CAAAUUGAAAUGUGGAAAAGGCGCUUAAUGCGCAUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGCAGCCAUCUGCGGACUUCUGCUCCAGCUUUUCCAACUGCUC ............((((((((((((.....))))...........((((((.(((((......))))))))).)).(((((((...((....))...)))))))..))))))))....... (-29.01 = -31.45 + 2.44)



| Location | 15,750,142 – 15,750,262 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -35.42 |
| Energy contribution | -35.05 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.98 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15750142 120 - 23771897 GCCAAAAGGCACAAAUGCGCCGUGCAGUCAGUUCGCUCGGCAAAUUGAAAUGUGGAAAAGGCGCUUAAUGCGCAUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGC (((....)))......((.(((.(((((((.((((.((.(((........))).))....((((.....))))...))))...)))...(((((((......))))))))))).))).)) ( -35.80) >DroSec_CAF1 1846 120 - 1 GCCAAAAGGCACAAAUGCGCCGUGCAGUCAGCUCGCUCGGCAAAUUGAAAUGUGGAAAAGGCGCUUAAUGCGCAUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGC (((....)))......((.(((.((((....(((..((.(((........))).))....((((.....))))............))).(((((((......))))))))))).))).)) ( -36.10) >DroSim_CAF1 2077 120 - 1 GCCAAAAGGCACAAAUGCGCCGUGCAGUCAGCUCGCUCGGCAAAUUGAAAUGUGGAAAAGGCGCUUAAUGCGCAUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGC (((....)))......((.(((.((((....(((..((.(((........))).))....((((.....))))............))).(((((((......))))))))))).))).)) ( -36.10) >DroEre_CAF1 1820 120 - 1 GCCAAAAGGCACAAAUGCGCCGUGCAGUCAGCUCGCUCGGCAAAUUGAAAUGUGGAAAAGGCGCUUAAUGUGCAUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGC .((.....(((((...((((((((.((....)))))((.(((........))).))...)))))....)))))...........((((((.(((((......))))))))).)).))... ( -36.00) >consensus GCCAAAAGGCACAAAUGCGCCGUGCAGUCAGCUCGCUCGGCAAAUUGAAAUGUGGAAAAGGCGCUUAAUGCGCAUAUGAAAAAUGGAGAAAGGAAAUUGCAUUUUCCUUCUGCCCGGAGC (((....)))......((.(((.((((....(((..((.(((........))).))....((((.....))))............))).(((((((......))))))))))).))).)) (-35.42 = -35.05 + -0.37)



| Location | 15,750,182 – 15,750,302 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -36.44 |
| Consensus MFE | -34.99 |
| Energy contribution | -34.99 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15750182 120 + 23771897 UUCAUAUGCGCAUUAAGCGCCUUUUCCACAUUUCAAUUUGCCGAGCGAACUGACUGCACGGCGCAUUUGUGCCUUUUGGCCUGGCCCACAACUAUUGCCACCUGGGCCUGUAAGCAUAUA ...((((((......((.(((.....((((....(((.(((((.(((.......))).))))).)))))))......)))))((((((..............)))))).....)))))). ( -34.74) >DroSec_CAF1 1886 120 + 1 UUCAUAUGCGCAUUAAGCGCCUUUUCCACAUUUCAAUUUGCCGAGCGAGCUGACUGCACGGCGCAUUUGUGCCUUUUGGCCUGGCCCACAACUAUUGCCACCUGGGCCGGUAAGCAUAUA ...((((((((.....))(((..................((((((...((.....))..(((((....))))).))))))..((((((..............)))))))))..)))))). ( -38.14) >DroSim_CAF1 2117 120 + 1 UUCAUAUGCGCAUUAAGCGCCUUUUCCACAUUUCAAUUUGCCGAGCGAGCUGACUGCACGGCGCAUUUGUGCCUUUUGGCCUGGCCCACAACUAUUGCCACCUGGGCCGGUAAGCAUAUA ...((((((((.....))(((..................((((((...((.....))..(((((....))))).))))))..((((((..............)))))))))..)))))). ( -38.14) >DroEre_CAF1 1860 120 + 1 UUCAUAUGCACAUUAAGCGCCUUUUCCACAUUUCAAUUUGCCGAGCGAGCUGACUGCACGGCGCAUUUGUGCCUUUUGGCCUGGCCCACAACUAUUGCCACCUGGGCCUGUAAGCAUAUA ...((((((......((.(((.....((((....(((.(((((.(((.......))).))))).)))))))......)))))((((((..............)))))).....)))))). ( -34.74) >consensus UUCAUAUGCGCAUUAAGCGCCUUUUCCACAUUUCAAUUUGCCGAGCGAGCUGACUGCACGGCGCAUUUGUGCCUUUUGGCCUGGCCCACAACUAUUGCCACCUGGGCCGGUAAGCAUAUA ...((((((......((.(((.....((((....(((.(((((.(((.......))).))))).)))))))......)))))((((((..............)))))).....)))))). (-34.99 = -34.99 + -0.00)

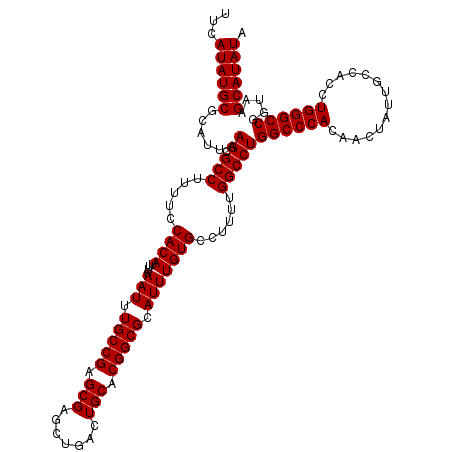
| Location | 15,750,182 – 15,750,302 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -40.04 |
| Energy contribution | -39.85 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15750182 120 - 23771897 UAUAUGCUUACAGGCCCAGGUGGCAAUAGUUGUGGGCCAGGCCAAAAGGCACAAAUGCGCCGUGCAGUCAGUUCGCUCGGCAAAUUGAAAUGUGGAAAAGGCGCUUAAUGCGCAUAUGAA (((((((.....(((((..(..((....))..))))))..(((....)))......(((((.(.((.((((((.((...)).))))))....)).)...))))).......))))))).. ( -39.90) >DroSec_CAF1 1886 120 - 1 UAUAUGCUUACCGGCCCAGGUGGCAAUAGUUGUGGGCCAGGCCAAAAGGCACAAAUGCGCCGUGCAGUCAGCUCGCUCGGCAAAUUGAAAUGUGGAAAAGGCGCUUAAUGCGCAUAUGAA (((((((.....(((((..(..((....))..))))))..(((....)))......((((((((.((....)))))((.(((........))).))...))))).......))))))).. ( -40.80) >DroSim_CAF1 2117 120 - 1 UAUAUGCUUACCGGCCCAGGUGGCAAUAGUUGUGGGCCAGGCCAAAAGGCACAAAUGCGCCGUGCAGUCAGCUCGCUCGGCAAAUUGAAAUGUGGAAAAGGCGCUUAAUGCGCAUAUGAA (((((((.....(((((..(..((....))..))))))..(((....)))......((((((((.((....)))))((.(((........))).))...))))).......))))))).. ( -40.80) >DroEre_CAF1 1860 120 - 1 UAUAUGCUUACAGGCCCAGGUGGCAAUAGUUGUGGGCCAGGCCAAAAGGCACAAAUGCGCCGUGCAGUCAGCUCGCUCGGCAAAUUGAAAUGUGGAAAAGGCGCUUAAUGUGCAUAUGAA (((((((.....(((((..(..((....))..))))))..(((....)))(((...((((((((.((....)))))((.(((........))).))...)))))....)))))))))).. ( -41.30) >consensus UAUAUGCUUACAGGCCCAGGUGGCAAUAGUUGUGGGCCAGGCCAAAAGGCACAAAUGCGCCGUGCAGUCAGCUCGCUCGGCAAAUUGAAAUGUGGAAAAGGCGCUUAAUGCGCAUAUGAA (((((((.....(((((..(..((....))..))))))..(((....)))......((((((((.((....)))))((.(((........))).))...))))).......))))))).. (-40.04 = -39.85 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:56 2006