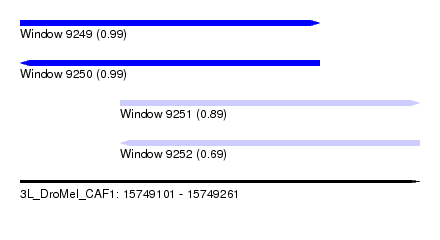
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,749,101 – 15,749,261 |
| Length | 160 |
| Max. P | 0.991409 |

| Location | 15,749,101 – 15,749,221 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -47.67 |
| Consensus MFE | -47.07 |
| Energy contribution | -47.30 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
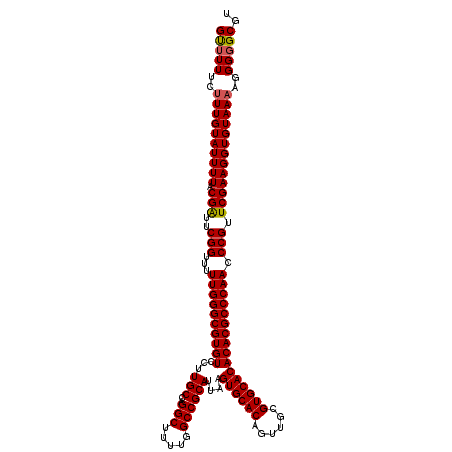
>3L_DroMel_CAF1 15749101 120 + 23771897 GUUUUUCUUUGUAUUUUACGAUUCGGUUUUUGGGCGUGUCCUUGCCAGGCUUUUGGCCGCAUUAAGUGCACAGUUGCGUGCACACACGCCCAACCCGUUCGAAGGUGUAAAAGGGGGCGU (((((..((((((((((.(((..(((...((((((((((...(((..(((.....))))))....((((((......)))))))))))))))).))).)))))))))))))..))))).. ( -48.60) >DroSec_CAF1 862 120 + 1 GUAUUUCUUUGUAUUUUACGAUUCGGUUUUUGGGCGUGUCCUUGCCAGGCUUUUGGCCGCAUUAAGUGCACAGUUGCGUGCACACACGCCCAACCCGUUCGAAGGUGUAAAAGGGGGCGU ((.(..((((..(((((.(((..(((...((((((((((...(((..(((.....))))))....((((((......)))))))))))))))).))).))))))))...))))..))).. ( -45.90) >DroEre_CAF1 873 119 + 1 GCUUUUCCUUGUAUUUUACGGUUCGGUUUUUGGGCGUGUCCUUGCCAGGCUU-UGGCCGCAUUAAGUGCACAGUUGCGUGCACACACGCCCAACCCGUUCGAAGGUGUAAAAGGGGGCGU (((..(((((.((((((.(((..(((...((((((((((...(((..(((..-..))))))....((((((......)))))))))))))))).))).)))))))))...)))))))).. ( -48.50) >consensus GUUUUUCUUUGUAUUUUACGAUUCGGUUUUUGGGCGUGUCCUUGCCAGGCUUUUGGCCGCAUUAAGUGCACAGUUGCGUGCACACACGCCCAACCCGUUCGAAGGUGUAAAAGGGGGCGU (((((..((((((((((.(((..(((...((((((((((...(((..(((.....))))))....((((((......)))))))))))))))).))).)))))))))))))..))))).. (-47.07 = -47.30 + 0.23)


| Location | 15,749,101 – 15,749,221 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -41.16 |
| Consensus MFE | -39.54 |
| Energy contribution | -39.32 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15749101 120 - 23771897 ACGCCCCCUUUUACACCUUCGAACGGGUUGGGCGUGUGUGCACGCAACUGUGCACUUAAUGCGGCCAAAAGCCUGGCAAGGACACGCCCAAAAACCGAAUCGUAAAAUACAAAGAAAAAC .......((((........(((.(((.(((((((((((((((((....)))))))........((((......))))....))))))))))...)))..)))........))))...... ( -41.09) >DroSec_CAF1 862 120 - 1 ACGCCCCCUUUUACACCUUCGAACGGGUUGGGCGUGUGUGCACGCAACUGUGCACUUAAUGCGGCCAAAAGCCUGGCAAGGACACGCCCAAAAACCGAAUCGUAAAAUACAAAGAAAUAC .......((((........(((.(((.(((((((((((((((((....)))))))........((((......))))....))))))))))...)))..)))........))))...... ( -41.09) >DroEre_CAF1 873 119 - 1 ACGCCCCCUUUUACACCUUCGAACGGGUUGGGCGUGUGUGCACGCAACUGUGCACUUAAUGCGGCCA-AAGCCUGGCAAGGACACGCCCAAAAACCGAACCGUAAAAUACAAGGAAAAGC ......((((.((......((..(((.(((((((((((((((((....)))))))........((((-.....))))....))))))))))...)))...)).....)).))))...... ( -41.30) >consensus ACGCCCCCUUUUACACCUUCGAACGGGUUGGGCGUGUGUGCACGCAACUGUGCACUUAAUGCGGCCAAAAGCCUGGCAAGGACACGCCCAAAAACCGAAUCGUAAAAUACAAAGAAAAAC .......((((........((..(((.(((((((((((((((((....)))))))........((((......))))....))))))))))...)))...))........))))...... (-39.54 = -39.32 + -0.22)

| Location | 15,749,141 – 15,749,261 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -54.23 |
| Consensus MFE | -52.53 |
| Energy contribution | -52.87 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
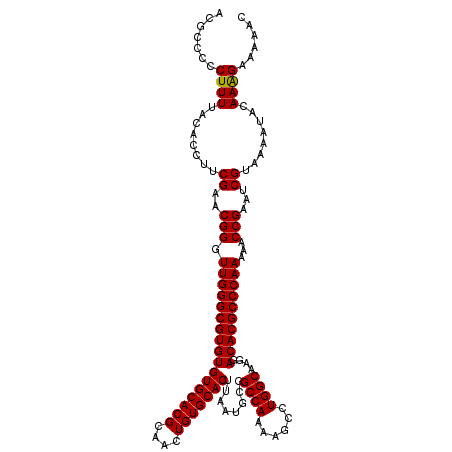
>3L_DroMel_CAF1 15749141 120 + 23771897 CUUGCCAGGCUUUUGGCCGCAUUAAGUGCACAGUUGCGUGCACACACGCCCAACCCGUUCGAAGGUGUAAAAGGGGGCGUGGCCGGUCGAAGGUGACGCCACGGGGCGCCCGGUAAGCAA ((((((.(((.(((((((((.....((((((......)))))).(((((((.((((.......)).))......)))))))).))))))))(....)(((....)))))).))))))... ( -55.20) >DroSec_CAF1 902 120 + 1 CUUGCCAGGCUUUUGGCCGCAUUAAGUGCACAGUUGCGUGCACACACGCCCAACCCGUUCGAAGGUGUAAAAGGGGGCGUGGCCGGUCGGAGGUGACGCCACGGGGCGCCCGGUAAGCAA ((((((.(((.(((((((((.....((((((......)))))).(((((((.((((.......)).))......)))))))).))))))))(....)(((....)))))).))))))... ( -54.80) >DroEre_CAF1 913 119 + 1 CUUGCCAGGCUU-UGGCCGCAUUAAGUGCACAGUUGCGUGCACACACGCCCAACCCGUUCGAAGGUGUAAAAGGGGGCGUGGCCGGUCGGAGGUGACGCCACGGGGCGCCCAGCAAGCAA (((((..(((((-(((((((.....((((((......)))))).(((((((.((((.......)).))......)))))))).))))))))(....)(((....))))))..)))))... ( -52.70) >consensus CUUGCCAGGCUUUUGGCCGCAUUAAGUGCACAGUUGCGUGCACACACGCCCAACCCGUUCGAAGGUGUAAAAGGGGGCGUGGCCGGUCGGAGGUGACGCCACGGGGCGCCCGGUAAGCAA ((((((.(((((((((((((.....((((((......)))))).(((((((.((((.......)).))......)))))))).))))))))).....(((....)))))).))))))... (-52.53 = -52.87 + 0.34)


| Location | 15,749,141 – 15,749,261 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -49.73 |
| Consensus MFE | -47.65 |
| Energy contribution | -47.43 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15749141 120 - 23771897 UUGCUUACCGGGCGCCCCGUGGCGUCACCUUCGACCGGCCACGCCCCCUUUUACACCUUCGAACGGGUUGGGCGUGUGUGCACGCAACUGUGCACUUAAUGCGGCCAAAAGCCUGGCAAG ...(((.(((((((((....)))(((......))).(((((((((((((.((.(......))).)))..))))))..(((((((....))))))).......))))....)))))).))) ( -48.40) >DroSec_CAF1 902 120 - 1 UUGCUUACCGGGCGCCCCGUGGCGUCACCUCCGACCGGCCACGCCCCCUUUUACACCUUCGAACGGGUUGGGCGUGUGUGCACGCAACUGUGCACUUAAUGCGGCCAAAAGCCUGGCAAG ...(((.(((((((((....)))(((......))).(((((((((((((.((.(......))).)))..))))))..(((((((....))))))).......))))....)))))).))) ( -48.40) >DroEre_CAF1 913 119 - 1 UUGCUUGCUGGGCGCCCCGUGGCGUCACCUCCGACCGGCCACGCCCCCUUUUACACCUUCGAACGGGUUGGGCGUGUGUGCACGCAACUGUGCACUUAAUGCGGCCA-AAGCCUGGCAAG ...(((((..((((((....)))(((......))).(((((((((((((.((.(......))).)))..))))))..(((((((....))))))).......)))).-..)))..))))) ( -52.40) >consensus UUGCUUACCGGGCGCCCCGUGGCGUCACCUCCGACCGGCCACGCCCCCUUUUACACCUUCGAACGGGUUGGGCGUGUGUGCACGCAACUGUGCACUUAAUGCGGCCAAAAGCCUGGCAAG ...(((.(((((((((....)))(((......))).(((((((((((((.((.(......))).)))..))))))..(((((((....))))))).......))))....)))))).))) (-47.65 = -47.43 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:49 2006