
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,748,882 – 15,749,021 |
| Length | 139 |
| Max. P | 0.615653 |

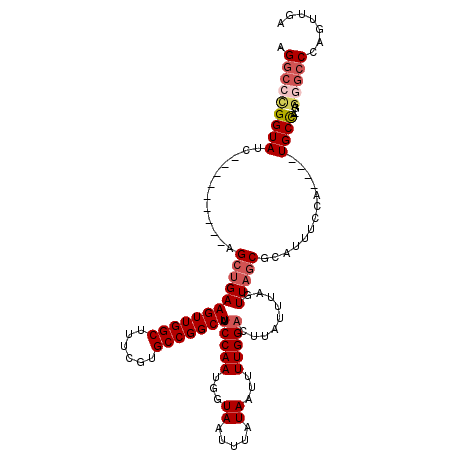
| Location | 15,748,882 – 15,748,981 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.99 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -22.02 |
| Energy contribution | -23.71 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15748882 99 - 23771897 AGGCUCGGUAUC---------AGCUGAAGUUGGCUUUCGUGCCGGCUAUCCAAUGGUAAUUUAUAAUUUUGGACUUAUUUAGUUAGCGCAUUUCCA----UGCU--------CCAGUUGA ..........((---------(((((.((((((.....((((..(((((((((...((.....))...)))))......))))..))))....)))----.)))--------.))))))) ( -26.20) >DroSec_CAF1 636 106 - 1 AGGCCCGGUAUC---------AGCUGAAGUUGGCUUUCGUGCCGGCUAUCCAAUGGUAAUUUAUAAUUUUGGACUUAUUUAGUUAGCGCAUUUCCA----UGCCA-GAGGGCCCAGUUGA .(((((......---------.((((((((((((......))))))).(((((...((.....))...))))).........)))))((((....)----)))..-..)))))....... ( -33.10) >DroSim_CAF1 691 106 - 1 AGGCCCGGUAUC---------AGCUGAAGUUGGCUUUCGUGCCGGCUAUCCAAUGGUAAUUUAUAAUUUUGGACUUAUUUAGUUAGCGCAUUUCCA----UGCCA-GAGGGCCAAGUUGA .(((((......---------.((((((((((((......))))))).(((((...((.....))...))))).........)))))((((....)----)))..-..)))))....... ( -34.00) >DroEre_CAF1 637 118 - 1 AGGCCUGGUAUCCUGGUAUCAGGAUGAAGUUGGCUUUCGUGCCGGCUAUCCAAUGGUAAUUUAUAAUUUUGG-CUUAU-UAGUUUGCGCAUUUCCACGGGUGCCAUGAGUGCCCAGCUGA .(((..((((..((((((((.((((..(((((((......)))))))))))((((((((....((((.....-...))-))..)))).))))......))))))).)..))))..))).. ( -37.90) >consensus AGGCCCGGUAUC_________AGCUGAAGUUGGCUUUCGUGCCGGCUAUCCAAUGGUAAUUUAUAAUUUUGGACUUAUUUAGUUAGCGCAUUUCCA____UGCCA_GAGGGCCCAGUUGA .(((((((((............((((((((((((......))))))).(((((...((.....))...))))).........))))).............))))....)))))....... (-22.02 = -23.71 + 1.69)


| Location | 15,748,910 – 15,749,021 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -19.17 |
| Energy contribution | -19.43 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15748910 111 + 23771897 AAAUAAGUCCAAAAUUAUAAAUUACCAUUGGAUAGCCGGCACGAAAGCCAACUUCAGCU---------GAUACCGAGCCUUCCCCAUAAUUUUGCACUUUUGCAGCUGUCCGAAAAAAGG ...........................(((((((((.(((.((..(((........)))---------.....)).))).............((((....)))))))))))))....... ( -23.80) >DroSec_CAF1 671 111 + 1 AAAUAAGUCCAAAAUUAUAAAUUACCAUUGGAUAGCCGGCACGAAAGCCAACUUCAGCU---------GAUACCGGGCCUUCCCCAUAAUUUUGCACUUUUGCAGCUGUCCGAAAAUAGG ...........................(((((((((.((...(((.(((....((....---------)).....))).))).)).......((((....)))))))))))))....... ( -24.30) >DroSim_CAF1 726 111 + 1 AAAUAAGUCCAAAAUUAUAAAUUACCAUUGGAUAGCCGGCACGAAAGCCAACUUCAGCU---------GAUACCGGGCCUUCCCCAUAAUUUUGCACUUUUGCAGCUGUCCGAAAAAAGG ...........................(((((((((.((...(((.(((....((....---------)).....))).))).)).......((((....)))))))))))))....... ( -24.30) >DroEre_CAF1 677 116 + 1 A-AUAAG-CCAAAAUUAUAAAUUACCAUUGGAUAGCCGGCACGAAAGCCAACUUCAUCCUGAUACCAGGAUACCAGGCCUUCCCCAUAAUUUUGAACUUUUGCAGCUGUCCGAAA--AGG .-.....-...................(((((((((..(((.(((.(((......((((((....))))))....))).)))..((......))......))).)))))))))..--... ( -28.30) >consensus AAAUAAGUCCAAAAUUAUAAAUUACCAUUGGAUAGCCGGCACGAAAGCCAACUUCAGCU_________GAUACCGGGCCUUCCCCAUAAUUUUGCACUUUUGCAGCUGUCCGAAAAAAGG ...........................(((((((((.(((......)))...........................................((((....)))))))))))))....... (-19.17 = -19.43 + 0.25)



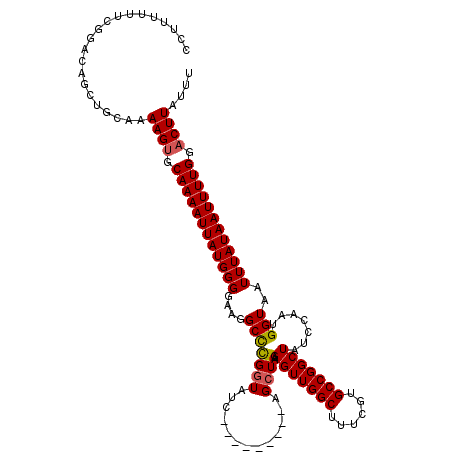
| Location | 15,748,910 – 15,749,021 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -25.62 |
| Energy contribution | -25.74 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15748910 111 - 23771897 CCUUUUUUCGGACAGCUGCAAAAGUGCAAAAUUAUGGGGAAGGCUCGGUAUC---------AGCUGAAGUUGGCUUUCGUGCCGGCUAUCCAAUGGUAAUUUAUAAUUUUGGACUUAUUU .....................((((.((((((((((.(((.(((.((((((.---------((((......))))...))))))))).)))(((....))))))))))))).)))).... ( -30.60) >DroSec_CAF1 671 111 - 1 CCUAUUUUCGGACAGCUGCAAAAGUGCAAAAUUAUGGGGAAGGCCCGGUAUC---------AGCUGAAGUUGGCUUUCGUGCCGGCUAUCCAAUGGUAAUUUAUAAUUUUGGACUUAUUU .....................((((.(((((((((((((....)))..((((---------(...(((((((((......))))))).))...)))))...)))))))))).)))).... ( -31.30) >DroSim_CAF1 726 111 - 1 CCUUUUUUCGGACAGCUGCAAAAGUGCAAAAUUAUGGGGAAGGCCCGGUAUC---------AGCUGAAGUUGGCUUUCGUGCCGGCUAUCCAAUGGUAAUUUAUAAUUUUGGACUUAUUU .....................((((.(((((((((((((....)))..((((---------(...(((((((((......))))))).))...)))))...)))))))))).)))).... ( -31.30) >DroEre_CAF1 677 116 - 1 CCU--UUUCGGACAGCUGCAAAAGUUCAAAAUUAUGGGGAAGGCCUGGUAUCCUGGUAUCAGGAUGAAGUUGGCUUUCGUGCCGGCUAUCCAAUGGUAAUUUAUAAUUUUGG-CUUAU-U .((--((((((....))).))))).(((((((((((((....(((((((((((((....))))))).(((((((......)))))))..)))..)))..)))))))))))))-.....-. ( -36.60) >consensus CCUUUUUUCGGACAGCUGCAAAAGUGCAAAAUUAUGGGGAAGGCCCGGUAUC_________AGCUGAAGUUGGCUUUCGUGCCGGCUAUCCAAUGGUAAUUUAUAAUUUUGGACUUAUUU .....................((((.((((((((((((....(((((((.............)))).(((((((......))))))).......)))..)))))))))))).)))).... (-25.62 = -25.74 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:46 2006