
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,748,481 – 15,748,631 |
| Length | 150 |
| Max. P | 0.939304 |

| Location | 15,748,481 – 15,748,591 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.29 |
| Mean single sequence MFE | -25.45 |
| Consensus MFE | -21.89 |
| Energy contribution | -22.95 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15748481 110 + 23771897 AUGGCCCAAAUGGCAUUUUGUCAUAAACGUUGUACUCGUUUUUGCCCUCUU----------UUUUUGCCACUCUUGUGUUGCCGACACUAGUGGCAUAUUUUCGUGCAAAAUUUCAUGUU ((((((.....)))(((((((((.(((((.......))))).)).......----------....(((((((...(((((...))))).))))))).........)))))))..)))... ( -24.80) >DroSec_CAF1 256 110 + 1 AUGGCCCAAAUGGAAUUUUGUCAUAAACGUUGUAGUCGUUGUUGGCCUCUU----------UUUUUGCCACUCUUGUGUUGCCGACACUAGUGGCAUAUUUUCGUGCAAAAUUUCACGUU ..........((((((((((.(((.......(..((((....))))..)..----------....(((((((...(((((...))))).))))))).......))))))))))))).... ( -27.70) >DroSim_CAF1 311 110 + 1 AUGGCCCAAAUGGAAUUUUGUCAUAAACGUUGUACUUGUUUUUGCCCUCUU----------UUUUUGCCACUCUUGUGUUGCCGACACUAGUGGCAUAUUUUCGUGCAAAAUUUCACGUU ..........(((((((((((((.(((((.......))))).)).......----------....(((((((...(((((...))))).))))))).........))))))))))).... ( -25.80) >DroEre_CAF1 252 119 + 1 AUGGCCCAAAUGGAAUUUUGUCAUAAACGUUGUAGUCGGUUUU-CCCUCUUUUUACUUUAUUUUUUGCCUCUGUUGAGUGGCCGACACUAGUGGCAUAUUUUCGUGCAAAAUUUCACGUU ..........((((((((((.(((....((...((..((....-))..))....)).........((((.((....((((.....)))))).)))).......))))))))))))).... ( -23.50) >consensus AUGGCCCAAAUGGAAUUUUGUCAUAAACGUUGUACUCGUUUUUGCCCUCUU__________UUUUUGCCACUCUUGUGUUGCCGACACUAGUGGCAUAUUUUCGUGCAAAAUUUCACGUU ..........((((((((((.((((((((.......)))))........................(((((((...((((.....)))).))))))).......))))))))))))).... (-21.89 = -22.95 + 1.06)

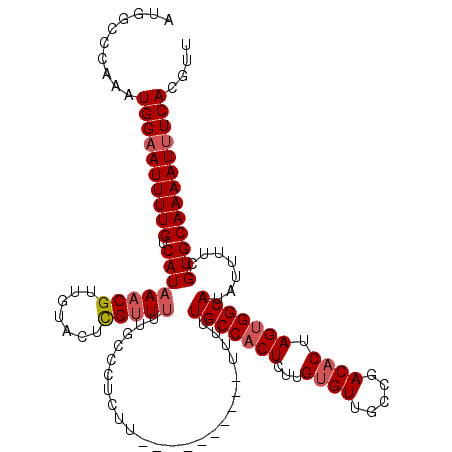
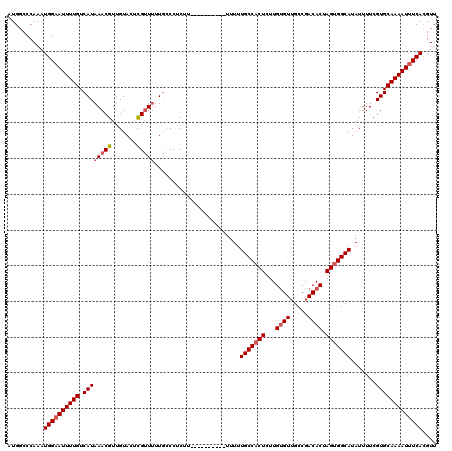
| Location | 15,748,481 – 15,748,591 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.29 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.60 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15748481 110 - 23771897 AACAUGAAAUUUUGCACGAAAAUAUGCCACUAGUGUCGGCAACACAAGAGUGGCAAAAA----------AAGAGGGCAAAAACGAGUACAACGUUUAUGACAAAAUGCCAUUUGGGCCAU ..(((((..((((((.........(((((((..(((.(....))))..)))))))....----------......))))))(((.......)))))))).......(((.....)))... ( -28.11) >DroSec_CAF1 256 110 - 1 AACGUGAAAUUUUGCACGAAAAUAUGCCACUAGUGUCGGCAACACAAGAGUGGCAAAAA----------AAGAGGCCAACAACGACUACAACGUUUAUGACAAAAUUCCAUUUGGGCCAU ..((((........))))......(((((((..(((.(....))))..)))))))....----------....((((...((((.......)))).....((((......)))))))).. ( -31.40) >DroSim_CAF1 311 110 - 1 AACGUGAAAUUUUGCACGAAAAUAUGCCACUAGUGUCGGCAACACAAGAGUGGCAAAAA----------AAGAGGGCAAAAACAAGUACAACGUUUAUGACAAAAUUCCAUUUGGGCCAU ..((((........))))......(((((((..(((.(....))))..)))))))....----------.....(((..((((.........))))...........((....))))).. ( -27.20) >DroEre_CAF1 252 119 - 1 AACGUGAAAUUUUGCACGAAAAUAUGCCACUAGUGUCGGCCACUCAACAGAGGCAAAAAAUAAAGUAAAAAGAGGG-AAAACCGACUACAACGUUUAUGACAAAAUUCCAUUUGGGCCAU ..((((........)))).......(((....(((((.(((.((....)).)))...................((.-....))))).))...........((((......)))))))... ( -20.20) >consensus AACGUGAAAUUUUGCACGAAAAUAUGCCACUAGUGUCGGCAACACAAGAGUGGCAAAAA__________AAGAGGGCAAAAACGACUACAACGUUUAUGACAAAAUUCCAUUUGGGCCAU ...(((.(((((((((..((....(((((((..(((.(....))))..)))))))..................(........)..........))..)).))))))).)))......... (-20.04 = -20.60 + 0.56)

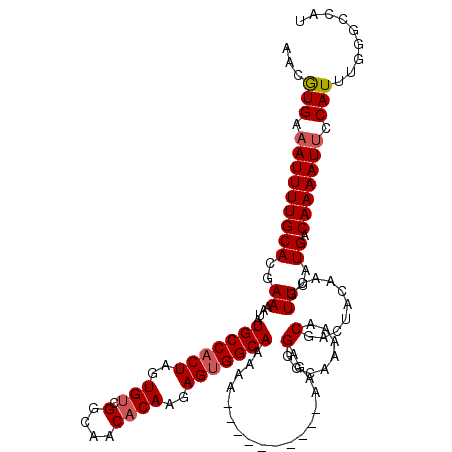
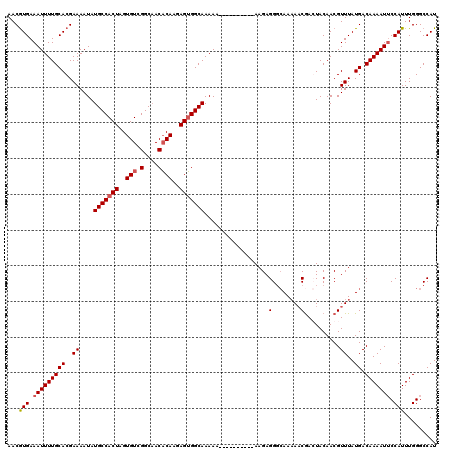
| Location | 15,748,521 – 15,748,631 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.70 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -20.48 |
| Energy contribution | -21.73 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15748521 110 - 23771897 UGUGAUUUUUCCCCAAAAACUCUCUCUUUAAAUAUGCAUGAACAUGAAAUUUUGCACGAAAAUAUGCCACUAGUGUCGGCAACACAAGAGUGGCAAAAA----------AAGAGGGCAAA .....................((((((((.....((((.((........)).))))........(((((((..(((.(....))))..)))))))...)----------))))))).... ( -27.80) >DroSec_CAF1 296 110 - 1 UGUGAUUUUUCCCCAAAAACUCUCUCUUUAAAUAUGCAUGAACGUGAAAUUUUGCACGAAAAUAUGCCACUAGUGUCGGCAACACAAGAGUGGCAAAAA----------AAGAGGCCAAC ......................(((((((.............((((........))))......(((((((..(((.(....))))..)))))))...)----------))))))..... ( -27.10) >DroSim_CAF1 351 110 - 1 UGUGAUUUUUCCCCAAAAACUCUCUCUUUAAAUAUGCAUGAACGUGAAAUUUUGCACGAAAAUAUGCCACUAGUGUCGGCAACACAAGAGUGGCAAAAA----------AAGAGGGCAAA .....................((((((((.............((((........))))......(((((((..(((.(....))))..)))))))...)----------))))))).... ( -29.60) >DroEre_CAF1 292 119 - 1 CGUGAUUCUGCUCCAAAAACGCUCCCUUUAAAUAUGCAAUAACGUGAAAUUUUGCACGAAAAUAUGCCACUAGUGUCGGCCACUCAACAGAGGCAAAAAAUAAAGUAAAAAGAGGG-AAA ......................(((((((...((((......))))...((((((..........(((((....)).)))..(((....)))))))))............))))))-).. ( -21.90) >consensus UGUGAUUUUUCCCCAAAAACUCUCUCUUUAAAUAUGCAUGAACGUGAAAUUUUGCACGAAAAUAUGCCACUAGUGUCGGCAACACAAGAGUGGCAAAAA__________AAGAGGGCAAA .....................(((((((..............((((........))))......(((((((..(((.(....))))..)))))))..............))))))).... (-20.48 = -21.73 + 1.25)

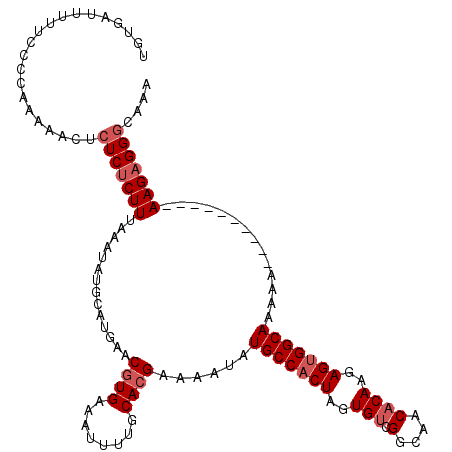
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:43 2006