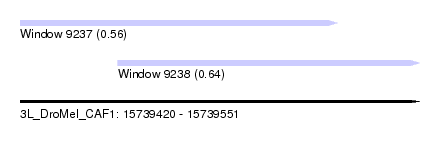
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,739,420 – 15,739,551 |
| Length | 131 |
| Max. P | 0.636545 |

| Location | 15,739,420 – 15,739,524 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 89.16 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -18.25 |
| Energy contribution | -18.65 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15739420 104 + 23771897 GAAAACUGAAGCUGCAACU------GGAGCUCAAAGGAAAUUAGCAAGCCAGAAAACAGAGCAGCCAAAUUUGGCAAAAGGAAAAUUCGUCGGCAGCUGGGGA--AACUUCU .........((((((..((------((.(((...........)))...))))...........((((....)))).................))))))(((..--..))).. ( -24.70) >DroSec_CAF1 54680 107 + 1 GAAAACUGAAGCUGCAACU-----UGGAGCUCAAAGGAAAUUAGCAAGCCCGAAAACAGAGCAGCCAAAUUUGGCAAAAGGAAAACUCCUCGGCAGCUGGGGAAAAACUUCA .....((..((((((....-----.(((((((...((............))(....).)))).((((....))))...........)))...))))))..)).......... ( -25.70) >DroSim_CAF1 56726 107 + 1 GAAAACUGAAGCUGCAACU-----UGGAGCUCAAAGGAAAUUAGCAAGCCCGAAAACAGAGCAGCCAAAUUUGGCAAAAGGAAAACUCCUCGGCAGCUGGGGAAAAACUUCA .....((..((((((....-----.(((((((...((............))(....).)))).((((....))))...........)))...))))))..)).......... ( -25.70) >DroEre_CAF1 46196 105 + 1 GAAAACUGAAGCUGGAACUGGAACUGGAGCUCAAAGGAAAUUAGCAAGCCAGAAAACAGAGCAGCCAAAUUUGGCAAA--GAAAAGUCUUCGGCAGCUGGGAA--AACU--- .....((..(((((...(((...((((.(((...........)))...))))....)))....((((....))))...--.............))))).))..--....--- ( -22.40) >DroYak_CAF1 155308 103 + 1 GAAAACUGAAGCUGGAGCUGG-------GCUCAAAGGAAAUUAGCAAGCUAGAAAACAGAGCAGCCAAAUUUGGCAAAAGGAAAAGUCUUCGGCAGCUGGGGA--AACUUCU ..........(((((((((..-------((((........((((....))))......)))).((((....)))).........)).)))))))....(((..--..))).. ( -24.74) >consensus GAAAACUGAAGCUGCAACU_____UGGAGCUCAAAGGAAAUUAGCAAGCCAGAAAACAGAGCAGCCAAAUUUGGCAAAAGGAAAACUCCUCGGCAGCUGGGGA__AACUUCA .....((..((((((.............((((..........................)))).((((....)))).................)))))).))........... (-18.25 = -18.65 + 0.40)



| Location | 15,739,452 – 15,739,551 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.35 |
| Mean single sequence MFE | -26.79 |
| Consensus MFE | -9.80 |
| Energy contribution | -11.42 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15739452 99 + 23771897 AAUUAGCAAGCCAGAAAACAGAGCAGC--CAAAUUUGGCAAAAGGAAAA---UUCGU-CGGCAGCUGGGGA--AACUUCUC---UGCUUCUGAAAAUGAGGCGAGUGCGA .....(((.(((.............((--((....))))..........---((((.-.(((((..(((..--..)))..)---))))..)))).....)))...))).. ( -28.90) >DroVir_CAF1 84767 94 + 1 AAUUAGCUAGCCAA-AAACGUAGUUGGUAUAAAUUUGA--UAG-AAAGAAACCGCCUGAAGGAAUUGGC-C--GACU---------CUAAUGAAAUGCAAGCGAAUGAGA .....(((.((...-......(((((((...(((((..--(((-...........)))...))))).))-)--))))---------..........)).)))........ ( -14.71) >DroSec_CAF1 54713 101 + 1 AAUUAGCAAGCCCGAAAACAGAGCAGC--CAAAUUUGGCAAAAGGAAAA---CUCCU-CGGCAGCUGGGGAAAAACUUCAC---UGCUGCUGAAAAUGAGGCGAGUGCGA .....(((.(((((...........((--((....))))....(((...---.)))(-((((((((((((.....))))).---.))))))))...)).)))...))).. ( -32.50) >DroSim_CAF1 56759 101 + 1 AAUUAGCAAGCCCGAAAACAGAGCAGC--CAAAUUUGGCAAAAGGAAAA---CUCCU-CGGCAGCUGGGGAAAAACUUCAC---UGCUGCUGAAAAUGAGGCGAGUGCGA .....(((.(((((...........((--((....))))....(((...---.)))(-((((((((((((.....))))).---.))))))))...)).)))...))).. ( -32.50) >DroEre_CAF1 46234 91 + 1 AAUUAGCAAGCCAGAAAACAGAGCAGC--CAAAUUUGGCAAA--GAAAA---GUCUU-CGGCAGCUGGGAA--AACU---------CUGCUGAAAAUAAGGCGAGUGCGA .....(((.(((.............((--((....))))...--.....---...((-((((((..((...--..))---------)))))))).....)))...))).. ( -26.00) >DroYak_CAF1 155339 102 + 1 AAUUAGCAAGCUAGAAAACAGAGCAGC--CAAAUUUGGCAAAAGGAAAA---GUCUU-CGGCAGCUGGGGA--AACUUCUCGGAUGCUGCUGAAAAUAAGGCGAGUACGA .....((..(((.............((--((....))))..........---...((-((((((((.((((--....)))).)..))))))))).....)))..)).... ( -26.10) >consensus AAUUAGCAAGCCAGAAAACAGAGCAGC__CAAAUUUGGCAAAAGGAAAA___CUCCU_CGGCAGCUGGGGA__AACUUC_C___UGCUGCUGAAAAUGAGGCGAGUGCGA .....(((.(((......((((...........)))).....................(((((((....................))))))).......)))...))).. ( -9.80 = -11.42 + 1.61)

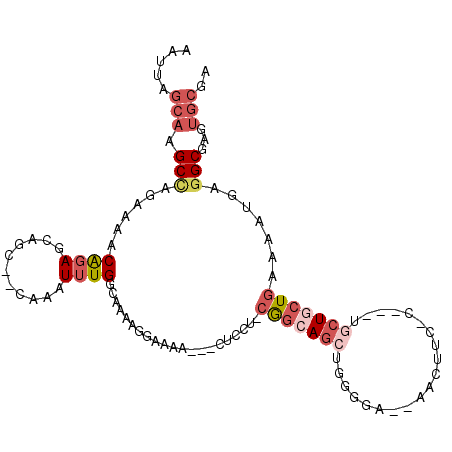

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:37 2006