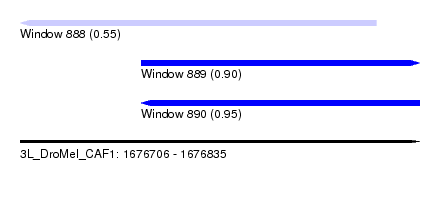
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,676,706 – 1,676,835 |
| Length | 129 |
| Max. P | 0.954157 |

| Location | 1,676,706 – 1,676,821 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.12 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -21.50 |
| Energy contribution | -22.17 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1676706 115 - 23771897 CAGUUGCUGCUGGGCCACGAAAUUGUUUU-GCGCAGCGGCAGAUGAGCUGUUGAAAUUGCAAGUGCGGCACUUGCUCAAUUUUUCAUGAGCCUCCCACUUCCACUUCUCGAUUUCA ((.(((((((((.((.(((....)))...-)).))))))))).)).(..(((((((.((.(((((.((.....(((((........)))))..))))))).)).)).)))))..). ( -34.40) >DroPse_CAF1 43674 94 - 1 GAGUU-----UGGGGCAAGAAAUUGGUCG-GCGCUGCGGCAGAUGAGCUGUUGAAAUUGCAAGUGCGGCACUUGCUCAAUUUUUCAUGAGCCUCCCACAA---------------- .....-----((((((((....)))...(-((((....))..(((((..(((((....(((((((...))))))))))))..)))))..))))))))...---------------- ( -30.00) >DroWil_CAF1 49334 95 - 1 AGGCU-----AAAGGAAAGAAAUGGUUUUGGCGCAGCGGCAGAUGAGCUGUUGAAAUUGCAAGUGCGGCACUUGCUCAAUUUUUCAUGAGCCUCCCACUA---------------- (((((-----....((((((.............(((((((......))))))).....(((((((...)))))))....))))))...))))).......---------------- ( -29.20) >DroMoj_CAF1 46063 97 - 1 --GCAGCUGCCAGGGAACGAGAUUGUUUU-GCGCAGCGGCAGAUGAGCUGUUGAAAUUGCAAGUGCGGCACUUGCUCAAUUUUUCAUGAACCUCCCACAA---------------- --((.(((((((..(((((....))))))-).))))).))..(((((..(((((....(((((((...))))))))))))..))))).............---------------- ( -29.00) >DroAna_CAF1 37511 99 - 1 AGGUUGCUGCCGGGGCAAGAAAUGGUUUU-GCGCAGCGGCAGAUGAGCUGUUGAAAUUGCAAGUGCGGCACUUGCUCAAUUUUUCAUGAGCCUCCCACAA---------------- .((..(((((((((((.(....).)))))-).)))))(((..(((((..(((((....(((((((...))))))))))))..)))))..)))..))....---------------- ( -36.70) >DroPer_CAF1 43813 94 - 1 GAGUU-----UGGGCCAAGAAAUUGGUCG-GCGCUGCGGCAGAUGAGCUGUUGAAAUUGCAAGUGCGGCACUUGCUCAAUUUUUCAUGAGCCUCCCACAA---------------- .(((.-----(.((((((....)))))).-).)))..(((..(((((..(((((....(((((((...))))))))))))..)))))..)))........---------------- ( -33.50) >consensus AAGUU_____UGGGGCAAGAAAUUGUUUU_GCGCAGCGGCAGAUGAGCUGUUGAAAUUGCAAGUGCGGCACUUGCUCAAUUUUUCAUGAGCCUCCCACAA________________ ...........(((................((...))(((..(((((..(((((....(((((((...))))))))))))..)))))..))).))).................... (-21.50 = -22.17 + 0.67)

| Location | 1,676,745 – 1,676,835 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.79 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.63 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1676745 90 + 23771897 GAGCAAGUGCCGCACUUGCAAUUUCAACAGCUCAUCUGCC---GCUGCGCAAAACAAUUUCGUGGCCCAGCAGCAACUGCAGUC---------------------UCAUUUUGA- ..(((((((...)))))))..........(((((..(((.---((((.((...((......)).)).)))).)))..)).))).---------------------.........- ( -24.60) >DroVir_CAF1 48472 101 + 1 GAGCAAGUGCCGCACUUGCAAUUUCAACAGCUCAUCUGCCGCCGCUGAGCAAAACAAUUUUGUUUCUUGGCAGCUGC--CAGU--------UUGAG-CCUCG---CCCGUUUUUU ..(((((((...)))))))..........(((((.((((.((.((..((..(((((....)))))))..)).)).).--))).--------.))))-)....---.......... ( -30.90) >DroGri_CAF1 45427 110 + 1 GAGCAAGUGCCGCACUUGCAAUUUCAACAGCUCAUCUGCC---GCUGCGCAAAACAAUUUCGUUCCUUGGCAGCUGC--CACUUUUGCAGCUUGGAGUCUCCUCUGCUGCUAAUG ..(((((((...)))))))..................((.---(((((.((((((......)))..)))))))).))--.......(((((..((.....))...)))))..... ( -32.80) >DroYak_CAF1 39448 90 + 1 GAGCAAGUGCCGCACUUGCAAUUUCAACAGCUCAUCUGCC---GCUGCGCAAAACAAUUUCGUGCCCCAGCAGCAGCUUCAGCC---------------------UCAUUUUCA- ..(((((((...)))))))..........(((...((((.---((((.(((...........)))..)))).))))....))).---------------------.........- ( -23.60) >DroMoj_CAF1 46086 105 + 1 GAGCAAGUGCCGCACUUGCAAUUUCAACAGCUCAUCUGCC---GCUGCGCAAAACAAUCUCGUUCCCUGGCAGCUGC--CAGU-UGGCAGUUUGGA-CUUUG---GGUGCUAUUU ..(((((((...)))))))..........(((((....((---(((((.((((((......)))..(((((....))--))))-)))))))..)).-...))---)))....... ( -32.10) >DroAna_CAF1 37534 89 + 1 GAGCAAGUGCCGCACUUGCAAUUUCAACAGCUCAUCUGCC---GCUGCGCAAAACCAUUUCUUGCCCCGGCAGCAACCUGGGCU---------------------UCGGAUUC-- ..(((((((...)))))))....((...((((((.(((((---(..(.((((.........))))).)))))).....))))))---------------------...))...-- ( -27.70) >consensus GAGCAAGUGCCGCACUUGCAAUUUCAACAGCUCAUCUGCC___GCUGCGCAAAACAAUUUCGUGCCCCGGCAGCAGC__CAGU______________________UCAGUUUCU_ ..(((((((...)))))))..........((......))....(((((.....((......))......)))))......................................... (-16.30 = -16.63 + 0.33)

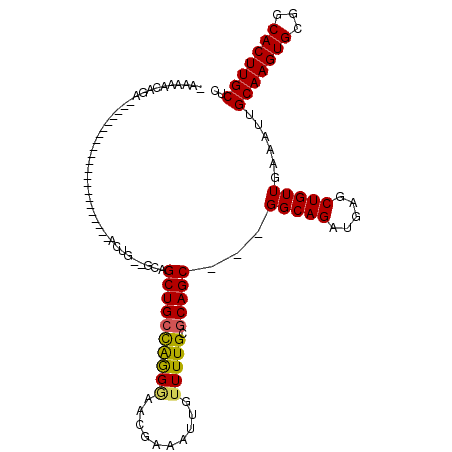
| Location | 1,676,745 – 1,676,835 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.79 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -18.35 |
| Energy contribution | -17.88 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1676745 90 - 23771897 -UCAAAAUGA---------------------GACUGCAGUUGCUGCUGGGCCACGAAAUUGUUUUGCGCAGC---GGCAGAUGAGCUGUUGAAAUUGCAAGUGCGGCACUUGCUC -((((....(---------------------(.((.((.(((((((((.((.(((....)))...)).))))---))))).)))))).))))....(((((((...))))))).. ( -30.40) >DroVir_CAF1 48472 101 - 1 AAAAAACGGG---CGAGG-CUCAA--------ACUG--GCAGCUGCCAAGAAACAAAAUUGUUUUGCUCAGCGGCGGCAGAUGAGCUGUUGAAAUUGCAAGUGCGGCACUUGCUC .......(((---(((((-((...--------(((.--(((((((....((((((....))))))...))))((((((......)))))).....))).)))..))).))))))) ( -34.90) >DroGri_CAF1 45427 110 - 1 CAUUAGCAGCAGAGGAGACUCCAAGCUGCAAAAGUG--GCAGCUGCCAAGGAACGAAAUUGUUUUGCGCAGC---GGCAGAUGAGCUGUUGAAAUUGCAAGUGCGGCACUUGCUC ..((((((((.(((....)))...(((((.....((--((....)))).((((((....))))))..)))))---.........))))))))....(((((((...))))))).. ( -41.80) >DroYak_CAF1 39448 90 - 1 -UGAAAAUGA---------------------GGCUGAAGCUGCUGCUGGGGCACGAAAUUGUUUUGCGCAGC---GGCAGAUGAGCUGUUGAAAUUGCAAGUGCGGCACUUGCUC -.........---------------------((((.(.(((((((((..((((......))))..).)))))---)))...).)))).........(((((((...))))))).. ( -31.10) >DroMoj_CAF1 46086 105 - 1 AAAUAGCACC---CAAAG-UCCAAACUGCCA-ACUG--GCAGCUGCCAGGGAACGAGAUUGUUUUGCGCAGC---GGCAGAUGAGCUGUUGAAAUUGCAAGUGCGGCACUUGCUC .((((((...---.....-......(((((.-...)--))))(((((..((((((....))))))((...))---)))))....))))))......(((((((...))))))).. ( -33.50) >DroAna_CAF1 37534 89 - 1 --GAAUCCGA---------------------AGCCCAGGUUGCUGCCGGGGCAAGAAAUGGUUUUGCGCAGC---GGCAGAUGAGCUGUUGAAAUUGCAAGUGCGGCACUUGCUC --...((...---------------------(((.((.((((((((((((((.(....).)))))).)))))---)))...)).)))...))....(((((((...))))))).. ( -32.50) >consensus _AAAAACAGA______________________ACUG__GCAGCUGCCAGGGAACGAAAUUGUUUUGCGCAGC___GGCAGAUGAGCUGUUGAAAUUGCAAGUGCGGCACUUGCUC .........................................((((((((((..........))))).)))))...(((((.....)))))......(((((((...))))))).. (-18.35 = -17.88 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:53 2006