
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,719,396 – 15,719,567 |
| Length | 171 |
| Max. P | 0.884088 |

| Location | 15,719,396 – 15,719,499 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 65.59 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -9.80 |
| Energy contribution | -10.05 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
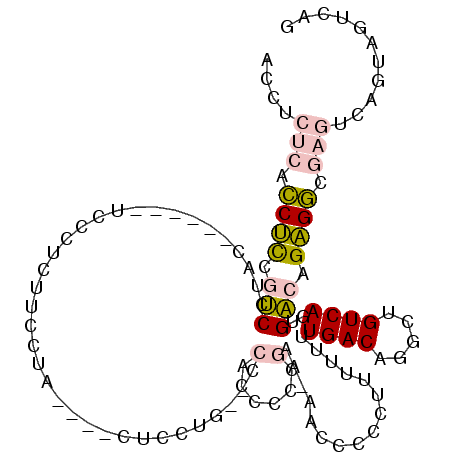
>3L_DroMel_CAF1 15719396 103 - 23771897 ACCUCUCACCUUCGUCCUU-------UCCCUCUUUCUACAAUCUCCUG--CCACCCUCGAA-AAACCCCUUUUUUUGACGGGCUGUCAGUGACAGAGGCGAGUCAGUAGUCAG ...................-------.....((..((((...(((..(--((.((((((((-(((.....)))))))).)))(((((...))))).))))))...))))..)) ( -24.60) >DroPse_CAF1 49872 99 - 1 AGCCCCUAGCCCUGUCCAAGUCGCUGUCCCUGUCCCAG----CUCCAUCUCCAUCUCC-----AUCUCUUUCUUUUGACAGCCUGUCACUGACAGGGC-----ACACACUCAG ........((((((((...(..((((.........)))----)..)............-----............((((.....))))..))))))))-----.......... ( -23.90) >DroSec_CAF1 34805 104 - 1 ACCUCUCACCUUCAUCCUCC------UCCCUCUUCCUACACACUCCUG--CCACACCCGAA-AACCCCCUUUUUUUGACGGGCUGUCAGUGACAGAGGCGAGUCAGUAGUCAG ....................------.....((..((((..((((..(--((...((((((-((........))))..))))(((((...))))).)))))))..))))..)) ( -20.50) >DroSim_CAF1 35897 95 - 1 ACCUCUCACCUUCAUCCUCU------CCCCUCUUCCUA------------CCACCCCCGAAAAACCCCCUUUUUUUGACGGGCUGUCAGUGACAGAGGCGAGUCAGUAGUCAG ..................((------(.(((((((((.------------.(((((.(((((((......)))))))..))).))..)).)).))))).)))........... ( -20.80) >DroYak_CAF1 71942 101 - 1 AGCUCUCACCUCCGCCCUAC--GC--CCCCCCUUCCUG----CUCCUG--CCGCCCCCGAA-AACC-CCCGUUUUUGACAGGCUGUCAGUGGCAGAGGCGAGUCAGUAGUGAG ....(((((..(.(..((.(--((--(........(((----(..(((--(.(((..((((-(((.-...)))))))...))).).)))..)))).)))))).).)..))))) ( -30.80) >DroPer_CAF1 49736 99 - 1 AGCCCCUAGCCCUGUCCAAGUCGCUGUCCCUGUCCCUG----UCCCAGCUCCAUCUCC-----AUCUCUUUCUUUUGACAGCCUGUCACUGACAGGGC-----ACACACUCAG ........((((((((...(..((((...(.......)----...))))..)......-----............((((.....))))..))))))))-----.......... ( -23.40) >consensus ACCUCUCACCUCCGUCCUAC______UCCCUCUUCCUA____CUCCUG__CCACCCCCGAA_AACCCCCUUUUUUUGACAGGCUGUCAGUGACAGAGGCGAGUCAGUAGUCAG ....(((.((((.(((...................................(......)................((((.....))))..))).)))).)))........... ( -9.80 = -10.05 + 0.25)

| Location | 15,719,466 – 15,719,567 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 65.46 |
| Mean single sequence MFE | -33.42 |
| Consensus MFE | -13.92 |
| Energy contribution | -15.73 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15719466 101 + 23771897 UGUAGAAAGAGGGA-------AAGGACGAAGGUGAGAGGUCCAAGAGGGGGAUUGAAUAUCCUCCAGUUGUCGUGAACGGCAAUGUGCCGCUAUUUUCUUUUAUCCUG ..........((((-------.((((.((((((.((.((..(....(((((((.....))))))).(((((((....))))))))..)).)))))))))))).)))). ( -35.00) >DroSec_CAF1 34875 102 + 1 UGUAGGAAGAGGGA------GGAGGAUGAAGGUGAGAGGUCCACAGAGGGGAUUGAAUGUCCUCCAGUUGUCGUGAACGACAAUGUGCCGCUAUCUUCUUGUAUUCUG ....((((...(.(------((((((((..(((............(.((((((.....))))))).(((((((....)))))))..)))..))))))))).).)))). ( -34.40) >DroSim_CAF1 35960 100 + 1 --UAGGAAGAGGGG------AGAGGAUGAAGGUGAGAGGUCCACAAAGGGGAUUGAAUGUCCUCCAGUUGUCGUGAACGGCAAUGCGCCGCUAUCUUCUUGUAUCCUG --(((((....(.(------((((((((..((((.............((((((.....)))).)).(((((((....))))))).))))..))))))))).).))))) ( -37.20) >DroEre_CAF1 26138 77 + 1 ------------------------------GGCAGGAGCAGGAGC-AGGCGAUUGAAUGUCCUCCAGUUGUCGUGAACGGUAAUGUGCCGCUAUCUGCUCUUAUCCUG ------------------------------((.((((((((((((-.((((((((.........))))))))......(((.....)))))).)))))))))..)).. ( -28.70) >DroYak_CAF1 72009 101 + 1 --CAGGAAGGGGGG--GC--GUAGGGCGGAGGUGAGAGCUACACG-AAGGGAUUGAAUGUCCUCCUGUUGUCGUGAACGGUAAUGUGCCGCUUUCUUCUCUUAUCCUG --((((((((((((--(.--..(((((((.(((....))).(((.-..(((((.....)))))...(((..((....))..))))))))))))))))))))).))))) ( -37.20) >DroPer_CAF1 49797 105 + 1 --CAGGGACAGGGACAGCGACUUGGACAGGGCUAGGGGCUAGGGG-CUGGAAGUAGAUGUCCUCCACUUUUUGUUAACUUUAAUGUUGCUCAUUGUGCCCCAUUGCAU --(..(..((((........))))..)..)((..(((((...(((-(.((((((.((.....)).))))))....(((......))))))).....)))))...)).. ( -28.00) >consensus __UAGGAAGAGGGA_______GAGGACGAAGGUGAGAGCUCCACG_AGGGGAUUGAAUGUCCUCCAGUUGUCGUGAACGGCAAUGUGCCGCUAUCUUCUCUUAUCCUG ..............................(((((((((........((((((.....))))))..(((((((....)))))))............))))))).)).. (-13.92 = -15.73 + 1.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:30 2006