| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,713,504 – 15,713,608 |
| Length | 104 |
| Max. P | 0.737922 |

| Location | 15,713,504 – 15,713,608 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 86.54 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -22.78 |
| Energy contribution | -22.45 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15713504 104 + 23771897 ACCAA-AAAAAGACAAUCCUAG-AGCUAAAGUUGGUUUACCGCUCGGCUGAGCGAAGAAGUUCAAUGCACAAGUUCAUGAACUUGGGGAAACCGACUUGAAAAG-GU .....-...........(((..-.....((((((((((.((((((....))))....(((((((..((....))...))))))).)).))))))))))....))-). ( -28.20) >DroSec_CAF1 28898 104 + 1 ACCAA-AAAAAGACAAUCCUGG-AGCUAAAGUUGGUUUACCGCUCGGCUGAGCGAAGAAGUUCAAUGCACAAGUUCAUGAACUUGGGGAAACCGACUUGAAAAG-GU .....-...........(((..-.....((((((((((.((((((....))))....(((((((..((....))...))))))).)).))))))))))....))-). ( -29.30) >DroSim_CAF1 30040 104 + 1 ACCGA-AAAAAGACAAUCCUGG-AGCUAAAGUUGGUUUACCGCUCGGCUGAGCGAAGAAGUUCAAUGCACAAGUUCAUGAACUUGGGGAAACCGACUUGAAAAG-GU .....-...........(((..-.....((((((((((.((((((....))))....(((((((..((....))...))))))).)).))))))))))....))-). ( -29.30) >DroYak_CAF1 65897 104 + 1 ACCGA-AAAAAGACAAUCCUAG-AGCUAAAGUUGGUUUACCGCUCGGCUGAGCGAAGAAGUUCAAUGCACAAGUUCAUGAACUUGGGGAGAGCGACUUGAAAAG-GU (((..-...(((.........(-(((((....))))))..(((((.(((((((......)))))..)).((((((....))))))....))))).))).....)-)) ( -27.50) >DroAna_CAF1 26629 102 + 1 ----A-AAAUAGACAAUCCAAGAAGCUAAAGUUGGUUUACCGCUCGGUUGAGCGAAGAAGUUCAAUGAUCAAGUUCAUGAACUUGGAGUAACCAACUUGAAGUGGGU ----.-.........(((((........(((((((((..((((((....))))....(((((((.(((......))))))))))))...)))))))))....))))) ( -31.00) >DroPer_CAF1 45357 101 + 1 GUCGAAAAAAAAACAAUCGUAA-AGUUUGAGUUGGUUUGCCGCUUGGCUAAGUGAAGAAGUUCAACGCACAAGUUCAUGAACUUGC----ACCAAUUCGUAAAG-GU ......................-.....((((((((..(((((((....)))))...(((((((((......))...)))))))))----))))))))......-.. ( -19.80) >consensus ACCGA_AAAAAGACAAUCCUAG_AGCUAAAGUUGGUUUACCGCUCGGCUGAGCGAAGAAGUUCAAUGCACAAGUUCAUGAACUUGGGGAAACCGACUUGAAAAG_GU ............................((((((((((..(((((....)))))...(((((((..((....))...)))))))....))))))))))......... (-22.78 = -22.45 + -0.33)

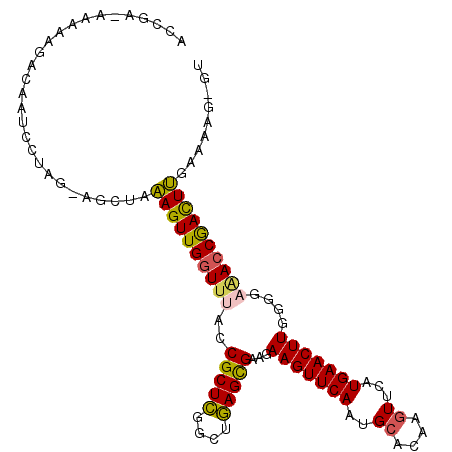
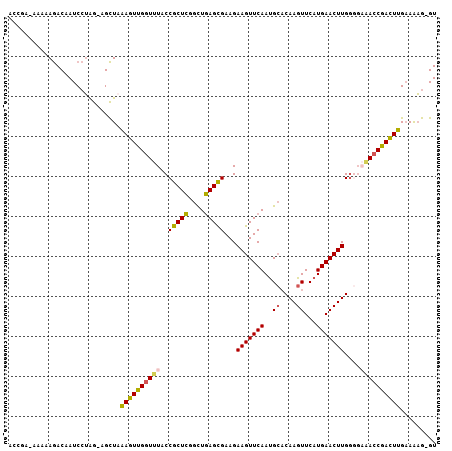
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:25 2006