
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
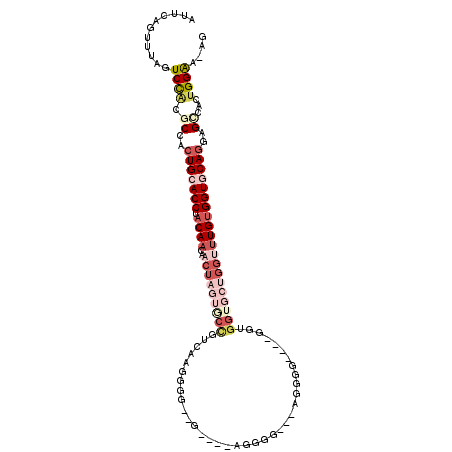
| Location | 15,708,762 – 15,708,868 |
| Length | 106 |
| Max. P | 0.736753 |

| Location | 15,708,762 – 15,708,868 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.39 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -18.52 |
| Energy contribution | -21.52 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
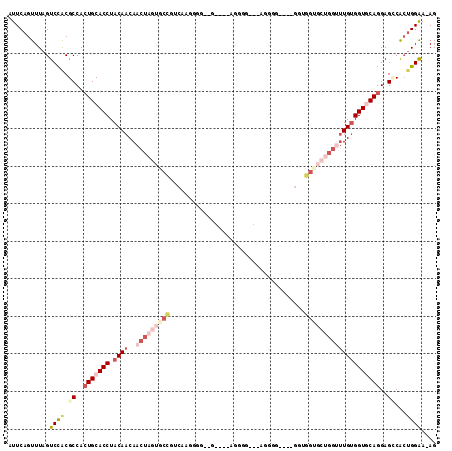
>3L_DroMel_CAF1 15708762 106 + 23771897 AUUCAGUUUAGUCCACGCCACUGCACCUACAACAACUAGUGCCGUCAAGUGGUAG----AUGGG---GGGGGUGGUGCUGGUGCUGGUUUGUGGUGCAGGAGCCACUGGAA-AG .((((((....(((.((((((.(((((..((...(((..(.(((((........)----)))).---)..)))..))..)))))......))))))..)))...)))))).-.. ( -37.50) >DroSec_CAF1 24288 84 + 1 AUUCAGUUUAGUCCGCGCCACUGCACCUACAACAACUAGUGCCGUCAAGU-----------------------------GGUGCUGGUUUGUGGUACAGGAGCCACUGGAA-AG .((((((...(....)((..(((.(((.((((..(((((..(((.....)-----------------------------))..)))))))))))).)))..)).)))))).-.. ( -28.90) >DroSim_CAF1 24744 102 + 1 AUUCAGUUUAGUCCACGCCACUGCACCUACAACAACUAGUACCGUCAAGUGGUUG----AUGGG---AGGGG----GGUGGUGCUGGAUUGUGGUACAGGAGCCACUGGAA-AG .((((((.((((((((((((((.(.(((.((.((((((.(.......).))))))----.))..---)))).----))))))).))))))).(((......))))))))).-.. ( -39.80) >DroEre_CAF1 15623 103 + 1 AUUCAGUUUAGUCCACGCCACUGCACCUACAACAACUC---CUGUCAAGGGGGGC----AGGGGCAGAGAGG----GGAGGUGCUGGUUUGUGGUGCAGGAGGCACUGGAAAAG .((((((.(..(((.((((((.((((((.(.....(((---(((((......)))----))))).......)----..))))))......))))))..))).).)))))).... ( -38.00) >DroYak_CAF1 61190 110 + 1 AUUCAGUUUAGUCUACGCCACUGCACCUACAACAACUAGUGCCGUCAAGGGGGGUGAAAGGGGGCCGAAGGG----GGUGGUGCUGGUUUGUGGUGCAGGAGGCACUGGAAAAG .((((((...(....)(((.(((((((.((((..(((((..((..(......(((........)))......----)..))..))))))))))))))))..))))))))).... ( -43.00) >DroAna_CAF1 21690 84 + 1 AUUCAGUUUAGUCCACGCCAGUGCACCUACAACAACUAGUCCUGUCGGGG----A----AGGGGC---------------------AACUGCGGUGCAGGAGUCGGCGGGA-AG ...........(((.((((..((((((........((..((((....)))----)----..))((---------------------....))))))))(....))))))))-.. ( -27.40) >consensus AUUCAGUUUAGUCCACGCCACUGCACCUACAACAACUAGUGCCGUCAAGGGG__G____AGGGG___AGGGG____GGUGGUGCUGGUUUGUGGUGCAGGAGCCACUGGAA_AG ...........((((.((..(((((((.((((..(((((((((....................................))))))))))))))))))))..))...)))).... (-18.52 = -21.52 + 3.00)

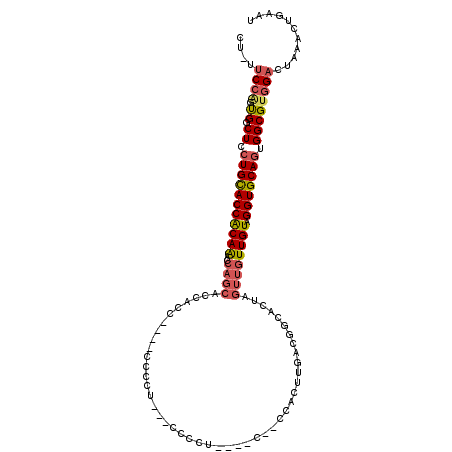
| Location | 15,708,762 – 15,708,868 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.39 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -16.76 |
| Energy contribution | -17.35 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
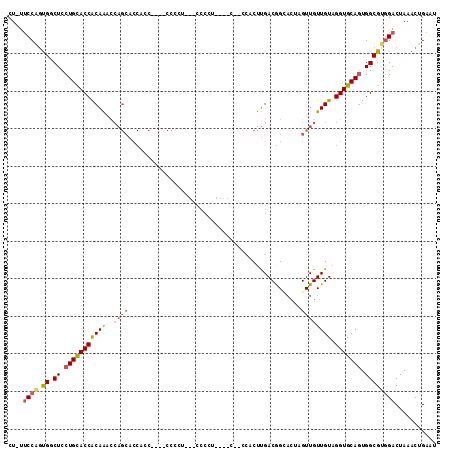
>3L_DroMel_CAF1 15708762 106 - 23771897 CU-UUCCAGUGGCUCCUGCACCACAAACCAGCACCAGCACCACCCCC---CCCAU----CUACCACUUGACGGCACUAGUUGUUGUAGGUGCAGUGGCGUGGACUAAACUGAAU ..-...((((.(((..(((...........)))..))).((((((.(---..(((----((((.....((((((....)))))))))))))..).)).)))).....))))... ( -29.20) >DroSec_CAF1 24288 84 - 1 CU-UUCCAGUGGCUCCUGUACCACAAACCAGCACC-----------------------------ACUUGACGGCACUAGUUGUUGUAGGUGCAGUGGCGCGGACUAAACUGAAU ..-...((((.....(((..((((......(((((-----------------------------...(((((((....)))))))..))))).))))..))).....))))... ( -26.20) >DroSim_CAF1 24744 102 - 1 CU-UUCCAGUGGCUCCUGUACCACAAUCCAGCACCACC----CCCCU---CCCAU----CAACCACUUGACGGUACUAGUUGUUGUAGGUGCAGUGGCGUGGACUAAACUGAAU ..-.((((...((..(((((((((((..((((......----.....---.((.(----(((....)))).)).....)))))))).)))))))..)).))))........... ( -24.33) >DroEre_CAF1 15623 103 - 1 CUUUUCCAGUGCCUCCUGCACCACAAACCAGCACCUCC----CCUCUCUGCCCCU----GCCCCCCUUGACAG---GAGUUGUUGUAGGUGCAGUGGCGUGGACUAAACUGAAU ....((((.((((..(((((((......(((((.((((----..((.........----.........))..)---))).)))))..))))))).))))))))........... ( -32.57) >DroYak_CAF1 61190 110 - 1 CUUUUCCAGUGCCUCCUGCACCACAAACCAGCACCACC----CCCUUCGGCCCCCUUUCACCCCCCUUGACGGCACUAGUUGUUGUAGGUGCAGUGGCGUAGACUAAACUGAAU ......((((.....((((.((((......(((((...----..(....)..................((((((....))))))...))))).)))).)))).....))))... ( -27.00) >DroAna_CAF1 21690 84 - 1 CU-UCCCGCCGACUCCUGCACCGCAGUU---------------------GCCCCU----U----CCCCGACAGGACUAGUUGUUGUAGGUGCACUGGCGUGGACUAAACUGAAU ..-((((((((.....((((((((((..---------------------((....----(----((......)))...))..)))).)))))).))))).)))........... ( -25.50) >consensus CU_UUCCAGUGGCUCCUGCACCACAAACCAGCACCACC____CCCCU___CCCCU____C__CCACUUGACGGCACUAGUUGUUGUAGGUGCAGUGGCGUGGACUAAACUGAAU ....((((.((.((.(((((((((((..((((..............................................)))))))).))))))).))))))))........... (-16.76 = -17.35 + 0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:23 2006