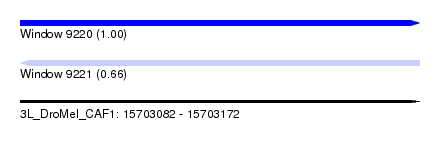
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,703,082 – 15,703,172 |
| Length | 90 |
| Max. P | 0.996744 |

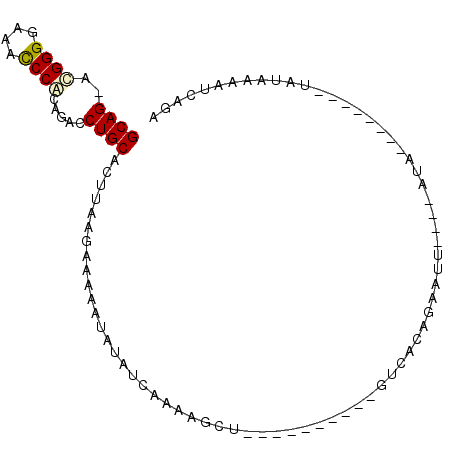
| Location | 15,703,082 – 15,703,172 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 68.83 |
| Mean single sequence MFE | -18.55 |
| Consensus MFE | -10.74 |
| Energy contribution | -10.92 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15703082 90 + 23771897 GCAGGGUGGGGAAACCCACAGACCUGCACUAAAGAAAAACAUCUCAAGAGCG----------GUCACAUAAUUGUAUAUAUGUACACUUAUGAAAUCAGA .....(((((....))))).((((.((.((..(((......)))..)).)))----------))).(((((.(((((....))))).)))))........ ( -30.20) >DroSec_CAF1 18609 91 + 1 GCAG-ACGGGGAAACCCACAGAACUGCACUUAAGAAAAAUAUAUCAAGAGCCAUAUAAUCUCGUCAUAUAAUUGUAUAUA--------UAUAAAAUCAGA ((((-..(((....)))......))))......((....(((((...(((.........)))...))))).(((((....--------)))))..))... ( -14.00) >DroEre_CAF1 9853 77 + 1 GCAG-ACGGGGAAACCCGCAGGCCUGCACUUAGGAAAAAUACACUCAAAGUU----------GUCACAGAUUU----AUA--------AAUAAAAUGGGA ((((-.((((....)))).....))))................((((...((----------((.........----...--------.))))..)))). ( -15.52) >DroYak_CAF1 55322 77 + 1 GCAG-ACGGGGAAAUCCACAAGCCUGCACUUAAGAAAAAUAUCUUAAAAGUU----------GCCAUAGAUUU----AUA--------UCUAUAAUCGGA ((((-.(..((....))....).))))..((((((......)))))).....----------...((((((..----..)--------)))))....... ( -14.50) >consensus GCAG_ACGGGGAAACCCACAGACCUGCACUUAAGAAAAAUAUAUCAAAAGCU__________GUCACAGAAUU____AUA________UAUAAAAUCAGA ((((..((((....)))).....))))......................................................................... (-10.74 = -10.92 + 0.19)

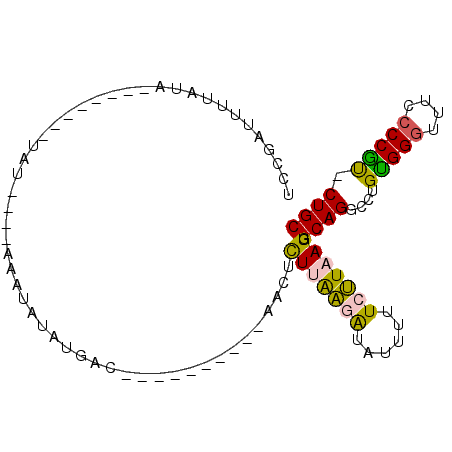
| Location | 15,703,082 – 15,703,172 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 68.83 |
| Mean single sequence MFE | -18.40 |
| Consensus MFE | -10.18 |
| Energy contribution | -9.80 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15703082 90 - 23771897 UCUGAUUUCAUAAGUGUACAUAUAUACAAUUAUGUGAC----------CGCUCUUGAGAUGUUUUUCUUUAGUGCAGGUCUGUGGGUUUCCCCACCCUGC ....((((((..((((..((((((......))))))..----------))))..)))))).............(((((...(((((....)))))))))) ( -28.70) >DroSec_CAF1 18609 91 - 1 UCUGAUUUUAUA--------UAUAUACAAUUAUAUGACGAGAUUAUAUGGCUCUUGAUAUAUUUUUCUUAAGUGCAGUUCUGUGGGUUUCCCCGU-CUGC .......(((((--------((........))))))).((((..(((((........)))))...))))....((((.....((((....)))).-)))) ( -13.60) >DroEre_CAF1 9853 77 - 1 UCCCAUUUUAUU--------UAU----AAAUCUGUGAC----------AACUUUGAGUGUAUUUUUCCUAAGUGCAGGCCUGCGGGUUUCCCCGU-CUGC ............--------...----....((((...----------.((((.(((.......)))..))))))))((..(((((....)))))-..)) ( -13.30) >DroYak_CAF1 55322 77 - 1 UCCGAUUAUAGA--------UAU----AAAUCUAUGGC----------AACUUUUAAGAUAUUUUUCUUAAGUGCAGGCUUGUGGAUUUCCCCGU-CUGC ...(.(((((((--------(..----..)))))))))----------....(((((((......))))))).((((((..(.((....))).))-)))) ( -18.00) >consensus UCCGAUUUUAUA________UAU____AAAUAUAUGAC__________AACUCUUAAGAUAUUUUUCUUAAGUGCAGGCCUGUGGGUUUCCCCGU_CUGC ....................................................(((((((......))))))).((((....(((((....))))).)))) (-10.18 = -9.80 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:21 2006