
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,696,834 – 15,697,076 |
| Length | 242 |
| Max. P | 0.956177 |

| Location | 15,696,834 – 15,696,942 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.01 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -20.32 |
| Energy contribution | -21.07 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
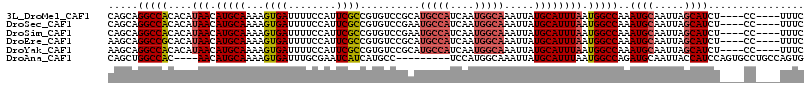
>3L_DroMel_CAF1 15696834 108 + 23771897 CAGCAGGCCACACAUAACAUGCAAAAGUGAUUUUCCAUUCGCCGUGUCCGCAUGCCAUCAAUGGCAAAUUAUGCAUUUAAUGGCCAAAUGCAAUUAGCAUCU----CC----UUUC .....(((((......(((((.....((((........)))))))))..((((((((....))))).....)))......)))))..((((.....))))..----..----.... ( -25.00) >DroSec_CAF1 12570 108 + 1 CAGCAGGCCACACAUAACAUGCAAAAGUGAUUUUCCAUUCGCCGUGUCCGAAUGCCAUCAAUGGCAAAUUAUGCAUUUAAUGGCCAAAUGCAAUUAGCAUCU----CC----UUUC .....(((((....(((.(((((...((((........))))..........(((((....))))).....)))))))).)))))..((((.....))))..----..----.... ( -24.70) >DroSim_CAF1 12502 108 + 1 CAGCAGGCCACACAUAACAUGCAAAAGUGAUUUUCCAUUCGCCGUGUCCGAAUGCCAUCAAUGGCAAAUUAUGCAUUUAAUGGCCAAAUGCAAUUAGCAUCU----CC----UUUC .....(((((....(((.(((((...((((........))))..........(((((....))))).....)))))))).)))))..((((.....))))..----..----.... ( -24.70) >DroEre_CAF1 3850 108 + 1 AAGCAGGCCGCACAUAACAUGCAAAAGUGAUUUUCCAUUCGCCGUGUCCGCAUGCCAUCAAUGGCAAAUUAUGCAUUUAAUGGCCAAAUGCAAUUAGCAUCU----CC----UUUC .....((((((((.....(((.((((....)))).))).....))))..((((((((....))))).....))).......))))..((((.....))))..----..----.... ( -24.80) >DroYak_CAF1 48594 108 + 1 AAGCAGGCCACACAUAACAUGCAAAAGUGAUUUUCCAUUCGCCGUGUCCGCAUGCCAUCAAUGGCAAAUUAUGCAUUUAAUGGCCAAAUGCAAUUAGCAUCU----CC----UUUC .....(((((......(((((.....((((........)))))))))..((((((((....))))).....)))......)))))..((((.....))))..----..----.... ( -25.00) >DroAna_CAF1 10671 103 + 1 CAGCUGGCCAC----AACAUGCAAAAGUGAUUUGCGAAUCAUCAUGCC---------UCCAUGGCAAAUUAUGCAUUUAAUGGCCAGAUGCAAUUACCAUCCAGUGCCUGCCAGUG ...(((((((.----...(((((...((((((....))))))..((((---------.....)))).....)))))....)))))))..(((..(((......)))..)))..... ( -29.90) >consensus CAGCAGGCCACACAUAACAUGCAAAAGUGAUUUUCCAUUCGCCGUGUCCGCAUGCCAUCAAUGGCAAAUUAUGCAUUUAAUGGCCAAAUGCAAUUAGCAUCU____CC____UUUC .....(((((....(((.(((((...((((........))))..........(((((....))))).....)))))))).)))))..((((.....))))................ (-20.32 = -21.07 + 0.75)

| Location | 15,696,874 – 15,696,979 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -34.55 |
| Consensus MFE | -22.93 |
| Energy contribution | -24.47 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
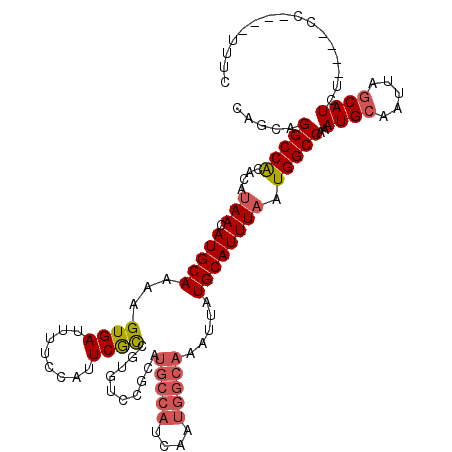
>3L_DroMel_CAF1 15696874 105 - 23771897 UUG---GGGGCUGGGAUCGUGGAAAGGACCUCUCUCCUUCGAAA----GG----AGAUGCUAAUUGCAUUUGGCCAUUAAAUGCAUAAUUUGCCAUUGAUGGCAUGCGGACACGGC ..(---((((..(((.((.(....).)))))..)))))((....----))----((((((.....)))))).((((((((..(((.....)))..))))))))...((....)).. ( -32.60) >DroSec_CAF1 12610 105 - 1 UUG---GGGGCUGGGGUCGUGGCAAGGACCUCUCUUCUUCGAAA----GG----AGAUGCUAAUUGCAUUUGGCCAUUAAAUGCAUAAUUUGCCAUUGAUGGCAUUCGGACACGGC ...---((((..((((((.(....).)))))).))))((((((.----..----((((((.....)))))).((((((((..(((.....)))..)))))))).))))))...... ( -35.30) >DroSim_CAF1 12542 104 - 1 -UG---GGGGCUGGGGUCGUGGAAAGGACCUCUCUCCUUCGAAA----GG----AGAUGCUAAUUGCAUUUGGCCAUUAAAUGCAUAAUUUGCCAUUGAUGGCAUUCGGACACGGC -..---((((..((((((.(....).)))))).))))((((((.----..----((((((.....)))))).((((((((..(((.....)))..)))))))).))))))...... ( -39.60) >DroEre_CAF1 3890 100 - 1 CUG---GGAGCUGGGG-----GAAAGGACCUCUCUCCUUUGAAA----GG----AGAUGCUAAUUGCAUUUGGCCAUUAAAUGCAUAAUUUGCCAUUGAUGGCAUGCGGACACGGC (((---..(((.((((-----.......))))((((((.....)----))----))).)))..((((((...((((((((..(((.....)))..))))))))))))))...))). ( -32.40) >DroYak_CAF1 48634 108 - 1 UUGAGGGGGGCUGGGGUCGUGAAAAGGACCUCUCUCCUGCGAAA----GG----AGAUGCUAAUUGCAUUUGGCCAUUAAAUGCAUAAUUUGCCAUUGAUGGCAUGCGGACACGGC .....(((((..((((((.(....).)))))).)))))(((...----..----((((((.....)))))).((((((((..(((.....)))..)))))))).)))......... ( -39.00) >DroAna_CAF1 10707 99 - 1 ------GUGGCCAGGACCUU--AAAGGACCUUUAUUCCACCACUGGCAGGCACUGGAUGGUAAUUGCAUCUGGCCAUUAAAUGCAUAAUUUGCCAUGGA---------GGCAUGAU ------...((((((.((..--...)).)))........(((.(((((((((...((((((...........))))))...))).....))))))))).---------)))..... ( -28.40) >consensus UUG___GGGGCUGGGGUCGUGGAAAGGACCUCUCUCCUUCGAAA____GG____AGAUGCUAAUUGCAUUUGGCCAUUAAAUGCAUAAUUUGCCAUUGAUGGCAUGCGGACACGGC ......((((..((((((.(....).)))))).)))).................((((((.....)))))).((((((((..(((.....)))..))))))))............. (-22.93 = -24.47 + 1.53)


| Location | 15,696,910 – 15,697,013 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.30 |
| Mean single sequence MFE | -18.58 |
| Consensus MFE | -15.08 |
| Energy contribution | -15.72 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
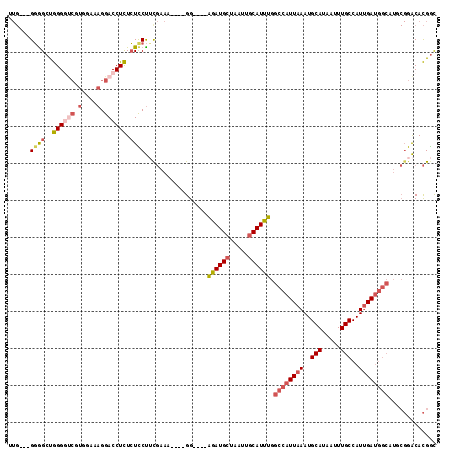
>3L_DroMel_CAF1 15696910 103 + 23771897 UUAAUGGCCAAAUGCAAUUAGCAUCUCCUUUCGAAGGAGAGAGGUCCUUUCCACGAUCCCAGCCCC---CAACCCACCAAUCCAAGCCCAUAUAGCAUA-----CCAUAGC ...((((....((((.((..((.((((((.....))))))(.((((........)))).)......---................))....)).)))).-----))))... ( -17.10) >DroSec_CAF1 12646 100 + 1 UUAAUGGCCAAAUGCAAUUAGCAUCUCCUUUCGAAGAAGAGAGGUCCUUGCCACGACCCCAGCCCC---CAACCC--------AACCCCAUAUAGCAUACCAUACCAUAGC ...((((....((((.....(((...((((((......))))))....)))...(....)......---......--------...........)))).))))........ ( -15.80) >DroSim_CAF1 12578 95 + 1 UUAAUGGCCAAAUGCAAUUAGCAUCUCCUUUCGAAGGAGAGAGGUCCUUUCCACGACCCCAGCCCC---CA-------------ACCCCAUAUAGCAUACCAUACCAUAGC ...((((....((((........((((((.....))))))(.((((........)))).)......---..-------------..........)))).))))........ ( -19.30) >DroEre_CAF1 3926 91 + 1 UUAAUGGCCAAAUGCAAUUAGCAUCUCCUUUCAAAGGAGAGAGGUCCUUUC-----CCCCAGCUCC---CAGCCC-------CAGCUCCAUGUAGCAUA-----CCACAGC .....((((...(((.....)))((((((.....))))))..)))).....-----.....((((.---.(((..-------..)))....).)))...-----....... ( -18.00) >DroYak_CAF1 48670 99 + 1 UUAAUGGCCAAAUGCAAUUAGCAUCUCCUUUCGCAGGAGAGAGGUCCUUUUCACGACCCCAGCCCCCCUCAACCC-------CCAUUCCAUAUUGCAUA-----CCAUUGC .((((((....(((((((.....((((((.....))))))(.((((........)))).)...............-------.........))))))).-----)))))). ( -22.70) >consensus UUAAUGGCCAAAUGCAAUUAGCAUCUCCUUUCGAAGGAGAGAGGUCCUUUCCACGACCCCAGCCCC___CAACCC_______CAACCCCAUAUAGCAUA_____CCAUAGC ...((((....((((........((((((.....))))))(.((((........)))).)..................................))))......))))... (-15.08 = -15.72 + 0.64)


| Location | 15,696,910 – 15,697,013 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.30 |
| Mean single sequence MFE | -30.81 |
| Consensus MFE | -21.22 |
| Energy contribution | -21.94 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
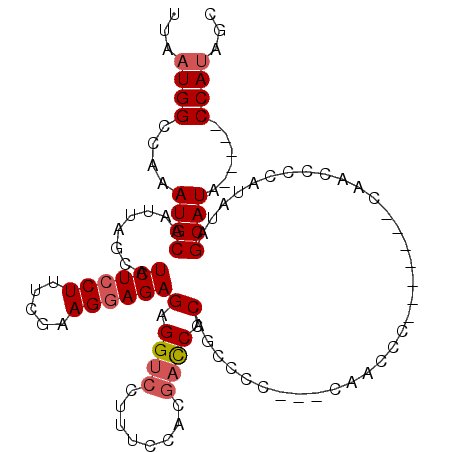
>3L_DroMel_CAF1 15696910 103 - 23771897 GCUAUGG-----UAUGCUAUAUGGGCUUGGAUUGGUGGGUUG---GGGGCUGGGAUCGUGGAAAGGACCUCUCUCCUUCGAAAGGAGAUGCUAAUUGCAUUUGGCCAUUAA ..(((((-----....)))))..(((((((((((((.....(---((((..(((.((.(....).))))))))))((((....))))..))))))).))...))))..... ( -28.20) >DroSec_CAF1 12646 100 - 1 GCUAUGGUAUGGUAUGCUAUAUGGGGUU--------GGGUUG---GGGGCUGGGGUCGUGGCAAGGACCUCUCUUCUUCGAAAGGAGAUGCUAAUUGCAUUUGGCCAUUAA ....(.((((((....)))))).)(((.--------.(((..---..(((.((((((.(....).))))))...(((((....))))).)))......)))..)))..... ( -31.80) >DroSim_CAF1 12578 95 - 1 GCUAUGGUAUGGUAUGCUAUAUGGGGU-------------UG---GGGGCUGGGGUCGUGGAAAGGACCUCUCUCCUUCGAAAGGAGAUGCUAAUUGCAUUUGGCCAUUAA ....(.((((((....)))))).)(((-------------..---((.((.((((((.(....).))))))((((((.....))))))........)).))..)))..... ( -32.50) >DroEre_CAF1 3926 91 - 1 GCUGUGG-----UAUGCUACAUGGAGCUG-------GGGCUG---GGAGCUGGGG-----GAAAGGACCUCUCUCCUUUGAAAGGAGAUGCUAAUUGCAUUUGGCCAUUAA (((((((-----....))))....)))..-------.(((((---((((..((((-----.......)))).)))))........((((((.....))))))))))..... ( -27.90) >DroYak_CAF1 48670 99 - 1 GCAAUGG-----UAUGCAAUAUGGAAUGG-------GGGUUGAGGGGGGCUGGGGUCGUGAAAAGGACCUCUCUCCUGCGAAAGGAGAUGCUAAUUGCAUUUGGCCAUUAA ..(((((-----((((((((.(((.....-------...(((.((((((..((((((.(....).)))))))))))).))).........))))))))))...)))))).. ( -33.63) >consensus GCUAUGG_____UAUGCUAUAUGGGGUUG_______GGGUUG___GGGGCUGGGGUCGUGGAAAGGACCUCUCUCCUUCGAAAGGAGAUGCUAAUUGCAUUUGGCCAUUAA .(((((.............))))).......................((((((((((.(....).))))))((((((.....))))))(((.....)))...))))..... (-21.22 = -21.94 + 0.72)

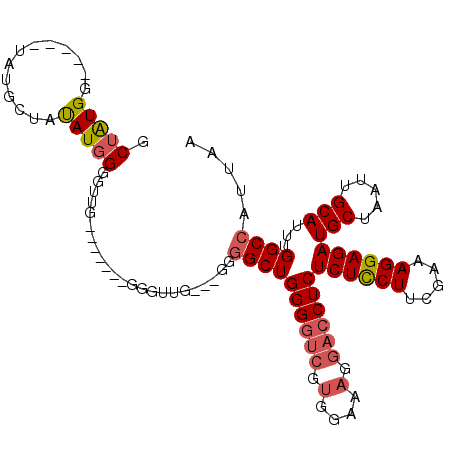
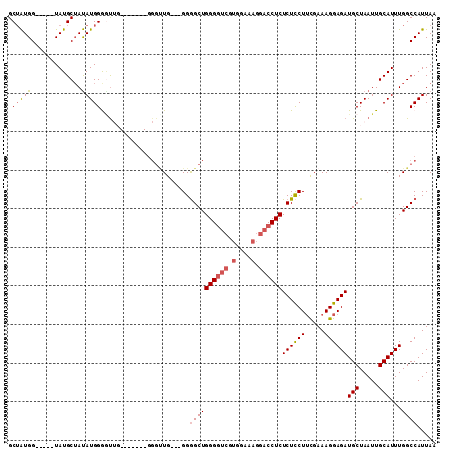
| Location | 15,696,979 – 15,697,076 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 76.21 |
| Mean single sequence MFE | -20.78 |
| Consensus MFE | -12.20 |
| Energy contribution | -12.87 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15696979 97 + 23771897 CCCACCAAUCCAAGCCCAUAUAGCAUA-----CCAUAGCAAUUGGGUGCGUGUCCAGCAUAAUUGCCCAAGAUUAACAUAAAUCAAUCAAAGGGCAUAAAGC .............(((((....((...-----.....))...)))))..((((((.(((....)))....((((......)))).......))))))..... ( -20.30) >DroSec_CAF1 12715 94 + 1 CCC--------AACCCCAUAUAGCAUACCAUACCAUAGCAAUUGGGUGCGUGUCCGGCAUAAUUGCCCAAGAUUAACAUAAAUCAAUCAAAGGCCAUAAAGC ...--------...........((((.(((............)))))))(((.((((((....))))...((((......)))).......)).)))..... ( -17.30) >DroSim_CAF1 12646 90 + 1 ------------ACCCCAUAUAGCAUACCAUACCAUAGCAAUUGGGUGCGUGUCCGGCAUAAUUGCCCAAGAUUAACAUAAAUCAAUCAAAGGCCAUAAAGC ------------..........((((.(((............)))))))(((.((((((....))))...((((......)))).......)).)))..... ( -17.30) >DroEre_CAF1 3990 90 + 1 CCC-------CAGCUCCAUGUAGCAUA-----CCACAGCAAUUGGGUGCGUGUCCGGCAUAAUUGCCCGAGAUUAACAUAAAUCAAUCAAAGGGCAUGAGGC ...-------..((..(((((.((((.-----(((.......)))))))....((((((....))))...((((......)))).......)))))))..)) ( -24.00) >DroYak_CAF1 48742 90 + 1 CCC-------CCAUUCCAUAUUGCAUA-----CCAUUGCAAUUGGGUGCGUGUCCGGCAUAAUUGCCCAAGAUUAACAUAAAUCAAUCAAAGGGCAUAAAGC ...-------.(((.(((.((((((..-----....)))))))))))).((((((((((....))))...((((......)))).......))))))..... ( -25.10) >DroPer_CAF1 32720 79 + 1 UCC-------CAGCUGCAUGGAAAACA-----UUACC-----------AGCUUCUGGCAUAAUUGCCAAAGAUUAAUAUAAAUCAAUCAGCGGGAAGAAAGU (((-------((((((.(((.....))-----)...)-----------))))..(((((....)))))..((((......)))).......))))....... ( -20.70) >consensus CCC_______CAACCCCAUAUAGCAUA_____CCAUAGCAAUUGGGUGCGUGUCCGGCAUAAUUGCCCAAGAUUAACAUAAAUCAAUCAAAGGGCAUAAAGC .....................................(((......)))((((((((((....))))...((((......)))).......))))))..... (-12.20 = -12.87 + 0.67)

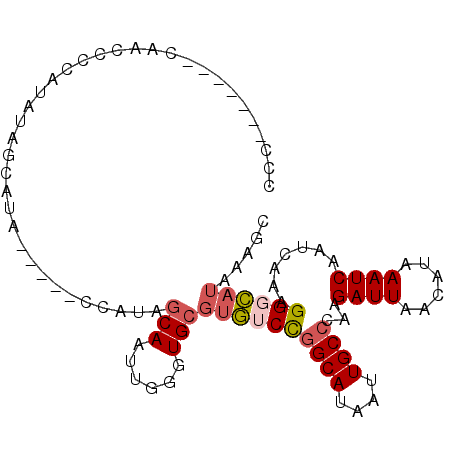
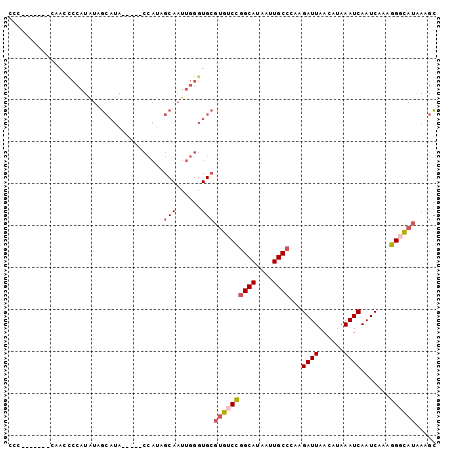
| Location | 15,696,979 – 15,697,076 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.21 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -11.92 |
| Energy contribution | -12.15 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15696979 97 - 23771897 GCUUUAUGCCCUUUGAUUGAUUUAUGUUAAUCUUGGGCAAUUAUGCUGGACACGCACCCAAUUGCUAUGG-----UAUGCUAUAUGGGCUUGGAUUGGUGGG ((....(((((...(((((((....)))))))..))))).....))....(((.((.((((.(.((((((-----....))))).).).))))..))))).. ( -26.90) >DroSec_CAF1 12715 94 - 1 GCUUUAUGGCCUUUGAUUGAUUUAUGUUAAUCUUGGGCAAUUAUGCCGGACACGCACCCAAUUGCUAUGGUAUGGUAUGCUAUAUGGGGUU--------GGG ((......((((..(((((((....)))))))..))))......))..........(((((((.(((((((((...)))))))).).))))--------))) ( -30.30) >DroSim_CAF1 12646 90 - 1 GCUUUAUGGCCUUUGAUUGAUUUAUGUUAAUCUUGGGCAAUUAUGCCGGACACGCACCCAAUUGCUAUGGUAUGGUAUGCUAUAUGGGGU------------ ((((((((((((..(((((((....)))))))..)))).(((((((((.....(((......)))..)))))))))......))))))))------------ ( -27.40) >DroEre_CAF1 3990 90 - 1 GCCUCAUGCCCUUUGAUUGAUUUAUGUUAAUCUCGGGCAAUUAUGCCGGACACGCACCCAAUUGCUGUGG-----UAUGCUACAUGGAGCUG-------GGG .((((((((((...(((((((....)))))))..)))))...(((((..(((.(((......))))))))-----)))(((......)))))-------))) ( -28.60) >DroYak_CAF1 48742 90 - 1 GCUUUAUGCCCUUUGAUUGAUUUAUGUUAAUCUUGGGCAAUUAUGCCGGACACGCACCCAAUUGCAAUGG-----UAUGCAAUAUGGAAUGG-------GGG ......(((((...(((((((....)))))))..)))))....(((.(....))))((((((((((....-----..))))))......)))-------).. ( -25.90) >DroPer_CAF1 32720 79 - 1 ACUUUCUUCCCGCUGAUUGAUUUAUAUUAAUCUUUGGCAAUUAUGCCAGAAGCU-----------GGUAA-----UGUUUUCCAUGCAGCUG-------GGA .......((((((((((((((....)))))))(((((((....))))))).(((-----------((...-----......))).))))).)-------))) ( -24.50) >consensus GCUUUAUGCCCUUUGAUUGAUUUAUGUUAAUCUUGGGCAAUUAUGCCGGACACGCACCCAAUUGCUAUGG_____UAUGCUAUAUGGAGUUG_______GGG ((((((((......(((((((....)))))))((.((((....)))).))................................))))))))............ (-11.92 = -12.15 + 0.23)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:17 2006