
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,696,328 – 15,696,484 |
| Length | 156 |
| Max. P | 0.784184 |

| Location | 15,696,328 – 15,696,444 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -26.76 |
| Energy contribution | -26.64 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15696328 116 + 23771897 AGACGCUUUUUCACAGUGGUAACUGCGGCACUAAGCAACUUUUCUCCACACCAUUUUUCUACCAGUCUAGAAAGUGCAGAUUUAAGUGCAGUUUCUGGAACAGCAAAUGGUUUUUG ....((((..((.((((....)))).))....)))).............(((((((..((.....((((((((.((((........)))).))))))))..)).)))))))..... ( -28.70) >DroSec_CAF1 12045 116 + 1 AGACGCUUUUUCACAGUGGUAACUGCGGCACUAAGCAACUUUUCUCCGCACCAUUUUUCUACCAGUCUAAAAAGUGCAGAUUUAAGUGCAGUUUCUGGAACAGCAAAUGGUUUUUG ....(((..(((.(((....((((((.((.....)).((((...((.((((..(((..(.....)...)))..)))).))...)))))))))).)))))).)))............ ( -24.10) >DroSim_CAF1 12004 116 + 1 AGACGCUUUUUCACAGUGGUAACUGCGGCACUAAGCAACUUUUCUCCGCACCAUUUUUCUGCCAGUCUAGAAAGUGCAGAUUUAAGUGCAGUUUCUGGAACAGCAAAUGGUUUUUG ....((((..((.((((....)))).))....)))).............((((((....(((...((((((((.((((........)))).))))))))...)))))))))..... ( -32.00) >DroEre_CAF1 3312 116 + 1 AGACGCUUUUUCACAGUGGUAAGUGCGGCACUAUGCAACUUUCCUCCAUGCCAUUUUUCUGCGAGUCUGGAAAGUGCAGAUUUAAGUGCAGUUUCUGGAGCAGCAAAUGGUUUUUA ...............((((.(((((((......))).))))....))))(((((((..((((...((..((((.((((........)))).))))..)))))).)))))))..... ( -33.40) >DroYak_CAF1 48032 116 + 1 AGACGCUUUUUCACAGUGGUAACUGCGCCACUAAGCAACUUUUCUCCACACCAUUUUUCUGCGAGUCUAGAAAGUGCAGAUUUAAGUGCAGUUUCUGGAGCAGCAAAUGGUUUUUG ....((((......((((((......)))))))))).............(((((((..((((...((((((((.((((........)))).)))))))))))).)))))))..... ( -35.80) >consensus AGACGCUUUUUCACAGUGGUAACUGCGGCACUAAGCAACUUUUCUCCACACCAUUUUUCUGCCAGUCUAGAAAGUGCAGAUUUAAGUGCAGUUUCUGGAACAGCAAAUGGUUUUUG ...............((((....(((........)))........))))(((((((..(((....((((((((.((((........)))).)))))))).))).)))))))..... (-26.76 = -26.64 + -0.12)

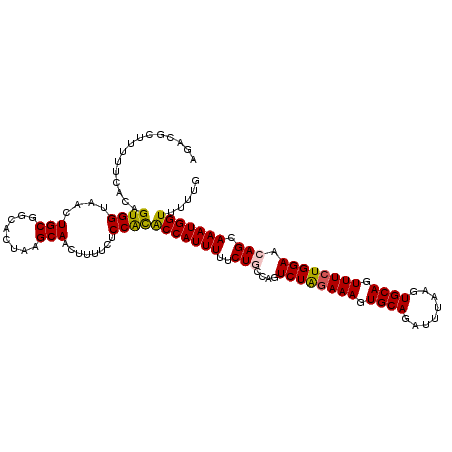
| Location | 15,696,328 – 15,696,444 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -24.12 |
| Energy contribution | -24.28 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15696328 116 - 23771897 CAAAAACCAUUUGCUGUUCCAGAAACUGCACUUAAAUCUGCACUUUCUAGACUGGUAGAAAAAUGGUGUGGAGAAAAGUUGCUUAGUGCCGCAGUUACCACUGUGAAAAAGCGUCU ............(((.((((((.((((((((((...((..(((((((((......)))))....))))..))...)))).((.....)).))))))....))).)))..))).... ( -28.60) >DroSec_CAF1 12045 116 - 1 CAAAAACCAUUUGCUGUUCCAGAAACUGCACUUAAAUCUGCACUUUUUAGACUGGUAGAAAAAUGGUGCGGAGAAAAGUUGCUUAGUGCCGCAGUUACCACUGUGAAAAAGCGUCU ............(((.((((((.((((((((((...(((((((((((((......)))))....))))))))...)))).((.....)).))))))....))).)))..))).... ( -28.50) >DroSim_CAF1 12004 116 - 1 CAAAAACCAUUUGCUGUUCCAGAAACUGCACUUAAAUCUGCACUUUCUAGACUGGCAGAAAAAUGGUGCGGAGAAAAGUUGCUUAGUGCCGCAGUUACCACUGUGAAAAAGCGUCU ............(((.((((((.((((((((((...((((((((((((........))))....))))))))...)))).((.....)).))))))....))).)))..))).... ( -29.70) >DroEre_CAF1 3312 116 - 1 UAAAAACCAUUUGCUGCUCCAGAAACUGCACUUAAAUCUGCACUUUCCAGACUCGCAGAAAAAUGGCAUGGAGGAAAGUUGCAUAGUGCCGCACUUACCACUGUGAAAAAGCGUCU ......((((((.((((((..((((.((((........)))).))))..))...))))..))))))(((((.((.((((.((........)))))).)).)))))........... ( -27.80) >DroYak_CAF1 48032 116 - 1 CAAAAACCAUUUGCUGCUCCAGAAACUGCACUUAAAUCUGCACUUUCUAGACUCGCAGAAAAAUGGUGUGGAGAAAAGUUGCUUAGUGGCGCAGUUACCACUGUGAAAAAGCGUCU ((...(((((((.((((((.(((((.((((........)))).))))).))...))))..))))))).))......((.(((((.....((((((....))))))...))))).)) ( -32.00) >consensus CAAAAACCAUUUGCUGUUCCAGAAACUGCACUUAAAUCUGCACUUUCUAGACUGGCAGAAAAAUGGUGUGGAGAAAAGUUGCUUAGUGCCGCAGUUACCACUGUGAAAAAGCGUCU .....(((((((.((((((.(((((.((((........)))).))))).))...))))..))))))).........((.(((((.....((((((....))))))...))))).)) (-24.12 = -24.28 + 0.16)



| Location | 15,696,365 – 15,696,484 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.76 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -20.54 |
| Energy contribution | -20.14 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.525959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15696365 119 + 23771897 ACUUUUCUCCACACCAUUUUUCUACCAGUCUAGAAAGUGCAGAUUUAAGUGCAGUUUCUGGAACAGCAAAUGGUUUUUGAGGUCUGAACGAAUCUAGAAACUAUAAACUUCUUACACUC .....((((...(((((((..((.....((((((((.((((........)))).))))))))..)).)))))))....))))((((........))))..................... ( -23.80) >DroSec_CAF1 12082 119 + 1 ACUUUUCUCCGCACCAUUUUUCUACCAGUCUAAAAAGUGCAGAUUUAAGUGCAGUUUCUGGAACAGCAAAUGGUUUUUGCGGUCUAAACGAAUCUAGAAACUAUAAACUUCUUACACUC ((((...((.((((..(((..(.....)...)))..)))).))...))))..((((((((((...(((((.....)))))..((.....)).))))))))))................. ( -22.80) >DroSim_CAF1 12041 119 + 1 ACUUUUCUCCGCACCAUUUUUCUGCCAGUCUAGAAAGUGCAGAUUUAAGUGCAGUUUCUGGAACAGCAAAUGGUUUUUGCGGUCUAAACGAAUCUAGAAACUAUAAACUUCUUACACUC ........((((((((((....(((...((((((((.((((........)))).))))))))...)))))))))....))))((((........))))..................... ( -30.20) >DroEre_CAF1 3349 119 + 1 ACUUUCCUCCAUGCCAUUUUUCUGCGAGUCUGGAAAGUGCAGAUUUAAGUGCAGUUUCUGGAGCAGCAAAUGGUUUUUACGGCUUAAACGAAUCUGGAACCUAUUAACUUCUUGCACUC .......((((.(((((((..((((...((..((((.((((........)))).))))..)))))).))))))).....((.......))....))))..................... ( -29.20) >DroYak_CAF1 48069 119 + 1 ACUUUUCUCCACACCAUUUUUCUGCGAGUCUAGAAAGUGCAGAUUUAAGUGCAGUUUCUGGAGCAGCAAAUGGUUUUUGCGGCUUAAACGAAUCUGGGAACUAUAAACUUCCCACACUC ........((.((((((((..((((...((((((((.((((........)))).)))))))))))).)))))))....).))............((((((........))))))..... ( -32.60) >consensus ACUUUUCUCCACACCAUUUUUCUGCCAGUCUAGAAAGUGCAGAUUUAAGUGCAGUUUCUGGAACAGCAAAUGGUUUUUGCGGUCUAAACGAAUCUAGAAACUAUAAACUUCUUACACUC ............(((((((..(((....((((((((.((((........)))).)))))))).))).)))))))........((((........))))..................... (-20.54 = -20.14 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:12 2006