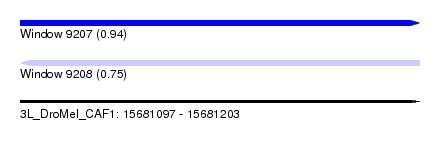
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,681,097 – 15,681,203 |
| Length | 106 |
| Max. P | 0.940539 |

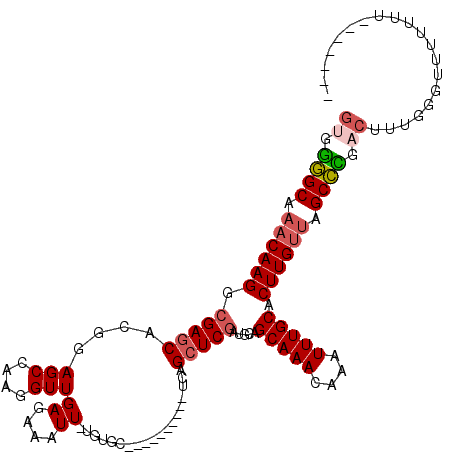
| Location | 15,681,097 – 15,681,203 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -27.50 |
| Energy contribution | -26.46 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15681097 106 + 23771897 GGAA---AAAAAACCCCAAAGUCGGGCUAACAAGUGCAAAUUUGUUUGCUGCAUCGAGCUGA----------GCACA-AAUUUCUCAACCUUGGCUCCGUGCUCGCCUUGUUUGCCCUAC ....---................((((.((((((.(((((....))))).((((.((((..(----------(....-...........))..)))).))))....)))))).))))... ( -32.66) >DroSec_CAF1 40578 102 + 1 -------AAAAAAACCCAAAGUCGGGCUAACAAGUGCAAAUUUGUUUGCUGCAUCGAGCUGA----------GCACA-AAUUUCUCAACCUUGGCUCCGUGCUCGCCUUGUUUGCCCCAC -------.............((.((((.((((((.(((((....))))).((((.((((..(----------(....-...........))..)))).))))....)))))).)))).)) ( -32.46) >DroSim_CAF1 40675 102 + 1 -------AAAAAAACCCAAAGUCGGGCUAACAAGUGCAAAUUUGUUUGCUGCAUCGAGCUGA----------GCACA-AAUUUCUCAACCUUGGCUCCGUGCUCGCCUUGUUUGCCCCAC -------.............((.((((.((((((.(((((....))))).((((.((((..(----------(....-...........))..)))).))))....)))))).)))).)) ( -32.46) >DroEre_CAF1 39863 107 + 1 AAAACCGAAAAAAGCC--AAGUCGGGCUGGCAAGUGCAAAUUUGUUUGCUGCAUCGAGCCGG----------GCACA-AAUUUCUCAACCUUGGCUCCGUGCUCGACUUGUUUGCCCCAC ................--..((.((((..(((((((((((....))))).....((((((((----------((.((-(...........))))).))).)))))))))))..)))).)) ( -36.40) >DroYak_CAF1 41206 109 + 1 GUGAGGUAAAAAAACCCAAAGUCAGGCUAACAAGUGCAAAUUUGUUUGCUGCAUCGAGCUGA----------GCACA-AAUUUCUCAACCUUGGCUCCGUGCUCGCCUUGUUUGCCCCGC .((.(((......)))))......(((.((((((.(((((....))))).((((.((((..(----------(....-...........))..)))).))))....)))))).))).... ( -31.56) >DroAna_CAF1 99258 104 + 1 ----------------CAAAGUUAAGCCAACAAGUGCAAAUUUGUUUGCUGCAUUGAGCAGAGAAGAGCAGGGCACAAAAUUUCUCAACCUUGGCUUCGUGUUCGCCUUGUUUGCUUCCU ----------------.......((((.((((((.((.....(((.....)))..(((((..((((..(((((...............))))).)))).))))))))))))).))))... ( -28.26) >consensus _______AAAAAAACCCAAAGUCGGGCUAACAAGUGCAAAUUUGUUUGCUGCAUCGAGCUGA__________GCACA_AAUUUCUCAACCUUGGCUCCGUGCUCGCCUUGUUUGCCCCAC ....................((.((((.((((((.(((((....))))).((((.(((((((............................))))))).))))....)))))).)))).)) (-27.50 = -26.46 + -1.05)

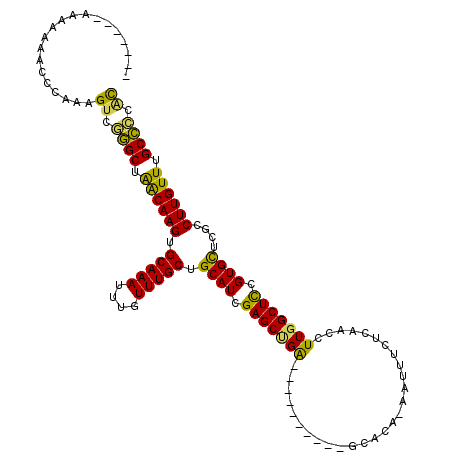
| Location | 15,681,097 – 15,681,203 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -24.87 |
| Energy contribution | -25.40 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15681097 106 - 23771897 GUAGGGCAAACAAGGCGAGCACGGAGCCAAGGUUGAGAAAUU-UGUGC----------UCAGCUCGAUGCAGCAAACAAAUUUGCACUUGUUAGCCCGACUUUGGGGUUUUUU---UUCC ((.((((.((((((.(((((...((((((((.........))-)).))----------)).))))).....(((((....))))).)))))).)))).))...((((......---)))) ( -36.80) >DroSec_CAF1 40578 102 - 1 GUGGGGCAAACAAGGCGAGCACGGAGCCAAGGUUGAGAAAUU-UGUGC----------UCAGCUCGAUGCAGCAAACAAAUUUGCACUUGUUAGCCCGACUUUGGGUUUUUUU------- ((.((((.((((((.(((((...((((((((.........))-)).))----------)).))))).....(((((....))))).)))))).)))).)).............------- ( -37.90) >DroSim_CAF1 40675 102 - 1 GUGGGGCAAACAAGGCGAGCACGGAGCCAAGGUUGAGAAAUU-UGUGC----------UCAGCUCGAUGCAGCAAACAAAUUUGCACUUGUUAGCCCGACUUUGGGUUUUUUU------- ((.((((.((((((.(((((...((((((((.........))-)).))----------)).))))).....(((((....))))).)))))).)))).)).............------- ( -37.90) >DroEre_CAF1 39863 107 - 1 GUGGGGCAAACAAGUCGAGCACGGAGCCAAGGUUGAGAAAUU-UGUGC----------CCGGCUCGAUGCAGCAAACAAAUUUGCACUUGCCAGCCCGACUU--GGCUUUUUUCGGUUUU ((.((((...((((((((((.(((.((((((.........))-)).))----------)))))))))....(((((....))))).))))...)))).))..--................ ( -36.40) >DroYak_CAF1 41206 109 - 1 GCGGGGCAAACAAGGCGAGCACGGAGCCAAGGUUGAGAAAUU-UGUGC----------UCAGCUCGAUGCAGCAAACAAAUUUGCACUUGUUAGCCUGACUUUGGGUUUUUUUACCUCAC ...((((.((((((.(((((...((((((((.........))-)).))----------)).))))).....(((((....))))).)))))).))))......((((......))))... ( -35.70) >DroAna_CAF1 99258 104 - 1 AGGAAGCAAACAAGGCGAACACGAAGCCAAGGUUGAGAAAUUUUGUGCCCUGCUCUUCUCUGCUCAAUGCAGCAAACAAAUUUGCACUUGUUGGCUUAACUUUG---------------- ...((((.(((((((((((...((((.((.(((.((....))....))).))..)))).((((.....))))........))))).)))))).)))).......---------------- ( -27.80) >consensus GUGGGGCAAACAAGGCGAGCACGGAGCCAAGGUUGAGAAAUU_UGUGC__________UCAGCUCGAUGCAGCAAACAAAUUUGCACUUGUUAGCCCGACUUUGGGUUUUUUU_______ ((.((((.((((((.(((((....(((....)))((....))...................))))).....(((((....))))).)))))).)))).)).................... (-24.87 = -25.40 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:00:09 2006