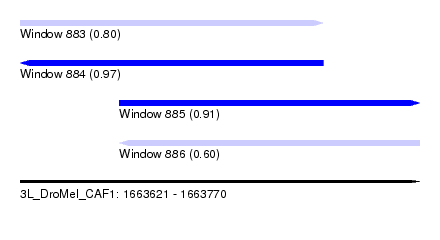
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,663,621 – 1,663,770 |
| Length | 149 |
| Max. P | 0.965176 |

| Location | 1,663,621 – 1,663,734 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.16 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1663621 113 + 23771897 AAAUUGCCAACAAGUUAUGAAAUUCCACAAAUUGUGAUUUGCAUGAAGCCGAAGCAGAAAUGGAAAAGGAACGA-GGAAAGG--AAAGGAAGUCCUGCAGGAAGCCCUGCAUUGGG ......((((............((((.....((((..(((.(((...((....))....))).)))....))))-.....))--))(((....)))(((((....))))).)))). ( -23.20) >DroSec_CAF1 25765 112 + 1 AAAUUGCCAACAAGUUAUGAAAUUCCACAAAUUGUGGUUUGCAUGAAGCCG-GGCAGAAAUGGAAAAGGGAACA-GGAAACG--AAAGGAAGUCCUGCAGGAAGUCCUGCAUUGGG ...(((((......(((((.....((((.....))))....))))).....-)))))..........(....)(-(((..(.--...)....))))(((((....)))))...... ( -32.30) >DroSim_CAF1 25856 96 + 1 AAAUUGCCAACAAGUUAUGAAAUUCCACAAAUUGUGAUUUGCAU-----------------GGAAAAGGGAACA-GGAAACG--AAAGGAAGUCCUGCAGGAAGCCCUGCAUUGGG ......((((...(((......((((((((((....)))))..)-----------------)))).....)))(-(((..(.--...)....))))(((((....))))).)))). ( -24.90) >DroEre_CAF1 29306 103 + 1 AAAUUGCCAGCAAGUUAUGGAAUUCCACAAAUUGUGAUUUGCAUGAAGGC-----GGCAGCAGAAAAG-GAACA-------GGAAGAGGAAGUCCUGCAGGAAGCCCUGCAUUGGG ...(((((.((...(((((.((...(((.....)))..)).)))))..))-----)))))(((.....-....(-------(((........))))(((((....))))).))).. ( -28.20) >DroYak_CAF1 26051 114 + 1 AAAUUGCCAACAAGUUAUGAAAUUCCACAAAUUGUGAUUUGCAUGAAGGCA-GGCAGAAGCAGAAAAA-GAACAAGAAAAAGGAAGAGGAAGUCCUGCAGGAAGCCCUGCACUGGG ...(((((..(...(((((((((..((.....))..)))).)))))..)..-)))))...(((.....-...........((((........))))(((((....))))).))).. ( -26.90) >consensus AAAUUGCCAACAAGUUAUGAAAUUCCACAAAUUGUGAUUUGCAUGAAGCC___GCAGAAACGGAAAAGGGAACA_GGAAACG__AAAGGAAGUCCUGCAGGAAGCCCUGCAUUGGG ......((((....(((((((((..((.....))..)))).)))))........................................(((....)))(((((....))))).)))). (-18.00 = -18.16 + 0.16)

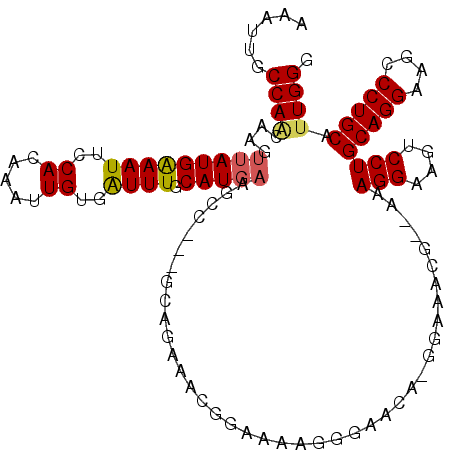
| Location | 1,663,621 – 1,663,734 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.41 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -18.66 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1663621 113 - 23771897 CCCAAUGCAGGGCUUCCUGCAGGACUUCCUUU--CCUUUCC-UCGUUCCUUUUCCAUUUCUGCUUCGGCUUCAUGCAAAUCACAAUUUGUGGAAUUUCAUAACUUGUUGGCAAUUU .((((((((((....))))).(((.((((...--.......-...................((....)).....((((((....)))))))))).))).......)))))...... ( -24.90) >DroSec_CAF1 25765 112 - 1 CCCAAUGCAGGACUUCCUGCAGGACUUCCUUU--CGUUUCC-UGUUCCCUUUUCCAUUUCUGCC-CGGCUUCAUGCAAACCACAAUUUGUGGAAUUUCAUAACUUGUUGGCAAUUU .....((((((....))))))((....))...--.......-..................((((-..((.....))...((((.....))))................)))).... ( -23.60) >DroSim_CAF1 25856 96 - 1 CCCAAUGCAGGGCUUCCUGCAGGACUUCCUUU--CGUUUCC-UGUUCCCUUUUCC-----------------AUGCAAAUCACAAUUUGUGGAAUUUCAUAACUUGUUGGCAAUUU .((((((((((....)))))(((....)))..--.(((...-.(((((.......-----------------..((((((....))))))))))).....)))..)))))...... ( -20.30) >DroEre_CAF1 29306 103 - 1 CCCAAUGCAGGGCUUCCUGCAGGACUUCCUCUUCC-------UGUUC-CUUUUCUGCUGCC-----GCCUUCAUGCAAAUCACAAUUUGUGGAAUUCCAUAACUUGCUGGCAAUUU ......((((((......((((((........)))-------)))..-...))))))((((-----((......))......(((.((((((....)))))).)))..)))).... ( -28.50) >DroYak_CAF1 26051 114 - 1 CCCAGUGCAGGGCUUCCUGCAGGACUUCCUCUUCCUUUUUCUUGUUC-UUUUUCUGCUUCUGCC-UGCCUUCAUGCAAAUCACAAUUUGUGGAAUUUCAUAACUUGUUGGCAAUUU ..(((((((((....))))))(((........)))............-.....)))....((((-(((......)))....((((.((((((....)))))).)))).)))).... ( -23.90) >consensus CCCAAUGCAGGGCUUCCUGCAGGACUUCCUUU__CGUUUCC_UGUUCCCUUUUCCACUUCUGC___GCCUUCAUGCAAAUCACAAUUUGUGGAAUUUCAUAACUUGUUGGCAAUUU .....((((((....))))))((....))............................................(((.....((((.((((((....)))))).))))..))).... (-18.66 = -18.70 + 0.04)

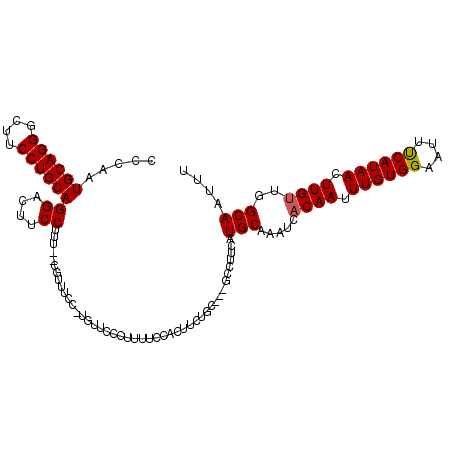
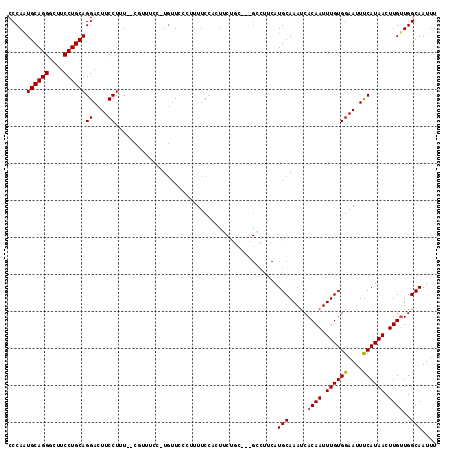
| Location | 1,663,658 – 1,663,770 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.08 |
| Mean single sequence MFE | -34.66 |
| Consensus MFE | -25.14 |
| Energy contribution | -25.34 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1663658 112 + 23771897 UUUGCAUGAAGCCGAAGCAGAAAUGGAAAAGGAACGA-GGAAAGG--AAAGGAAGUCCUGCAGGAAGCCCUGCAUUGGGCUUAAAGCUACGAGGGGCUCACUCGGCCAUGUCUUA ...(((((..(((((..((....))............-.......--...(..(((((((((((....))))).((((((.....))).)))))))))..))))))))))).... ( -33.40) >DroSec_CAF1 25802 111 + 1 UUUGCAUGAAGCCG-GGCAGAAAUGGAAAAGGGAACA-GGAAACG--AAAGGAAGUCCUGCAGGAAGUCCUGCAUUGGGCUUAAAGCUUCGAGGGGCUCACUCGGCCAUGUCUUA ...(((((..((((-((((....)).....(((..(.-(....)(--((...((((((((((((....))))))..)))))).....)))...)..))).))))))))))).... ( -39.30) >DroSim_CAF1 25893 95 + 1 UUUGCAU-----------------GGAAAAGGGAACA-GGAAACG--AAAGGAAGUCCUGCAGGAAGCCCUGCAUUGGGCUUAAAGCUACGAGGGGCUCACUCGGCCAUGUCUUA ...((((-----------------((....(((..((-(((..(.--...)....)))))...((.(((((......(((.....)))....))))))).)))..)))))).... ( -33.00) >DroEre_CAF1 29343 102 + 1 UUUGCAUGAAGGC-----GGCAGCAGAAAAG-GAACA-------GGAAGAGGAAGUCCUGCAGGAAGCCCUGCAUUGGGCUUAAAGCUACGAGGGGCCCACUCGGCCAUGUCUUA ........(((((-----((((((.......-...((-------(((........)))))....((((((......))))))...))).(((((....).)))))))..))))). ( -33.10) >DroYak_CAF1 26088 113 + 1 UUUGCAUGAAGGCA-GGCAGAAGCAGAAAAA-GAACAAGAAAAAGGAAGAGGAAGUCCUGCAGGAAGCCCUGCACUGGGCUUAAAGCUACGAGGAGCCCACUCGGCCAUGUCUUA ........((((((-(((...(((.......-...(........).......((((((((((((....))))))..))))))...))).((((.......))))))).)))))). ( -34.50) >consensus UUUGCAUGAAGCC___GCAGAAACGGAAAAGGGAACA_GGAAACG__AAAGGAAGUCCUGCAGGAAGCCCUGCAUUGGGCUUAAAGCUACGAGGGGCUCACUCGGCCAUGUCUUA ...(((((............................................((((((((((((....))))))..)))))).......((((.......))))..))))).... (-25.14 = -25.34 + 0.20)

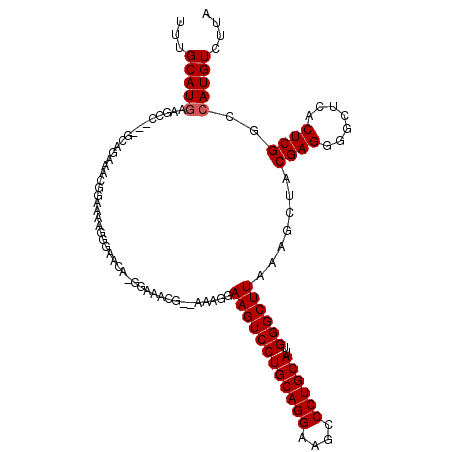
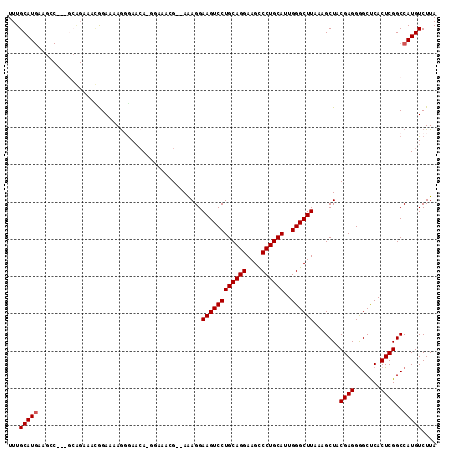
| Location | 1,663,658 – 1,663,770 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.08 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -18.42 |
| Energy contribution | -19.02 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1663658 112 - 23771897 UAAGACAUGGCCGAGUGAGCCCCUCGUAGCUUUAAGCCCAAUGCAGGGCUUCCUGCAGGACUUCCUUU--CCUUUCC-UCGUUCCUUUUCCAUUUCUGCUUCGGCUUCAUGCAAA ..(((.((((..(((.((((.....((((....((((((......)))))).))))((((.......)--)))....-..)))))))..)))).)))((....)).......... ( -31.30) >DroSec_CAF1 25802 111 - 1 UAAGACAUGGCCGAGUGAGCCCCUCGAAGCUUUAAGCCCAAUGCAGGACUUCCUGCAGGACUUCCUUU--CGUUUCC-UGUUCCCUUUUCCAUUUCUGCC-CGGCUUCAUGCAAA ..(((.((((..(((.((((....(((((....(((.((..((((((....)))))))).)))..)))--)).....-.)))).)))..)))).)))...-..((.....))... ( -29.00) >DroSim_CAF1 25893 95 - 1 UAAGACAUGGCCGAGUGAGCCCCUCGUAGCUUUAAGCCCAAUGCAGGGCUUCCUGCAGGACUUCCUUU--CGUUUCC-UGUUCCCUUUUCC-----------------AUGCAAA ...(.(((((..(((.((((.....((((....((((((......)))))).))))(((....)))..--.......-.)))).)))..))-----------------))))... ( -27.30) >DroEre_CAF1 29343 102 - 1 UAAGACAUGGCCGAGUGGGCCCCUCGUAGCUUUAAGCCCAAUGCAGGGCUUCCUGCAGGACUUCCUCUUCC-------UGUUC-CUUUUCUGCUGCC-----GCCUUCAUGCAAA ...((...(((.(((.(....))))(((((...((((((......))))))...((((((........)))-------)))..-.......))))).-----))).))....... ( -30.50) >DroYak_CAF1 26088 113 - 1 UAAGACAUGGCCGAGUGGGCUCCUCGUAGCUUUAAGCCCAGUGCAGGGCUUCCUGCAGGACUUCCUCUUCCUUUUUCUUGUUC-UUUUUCUGCUUCUGCC-UGCCUUCAUGCAAA (((((...((..(((..((.((((.((((....((((((......)))))).))))))))))..)))..))....)))))...-.......((....)).-(((......))).. ( -30.50) >consensus UAAGACAUGGCCGAGUGAGCCCCUCGUAGCUUUAAGCCCAAUGCAGGGCUUCCUGCAGGACUUCCUUU__CGUUUCC_UGUUCCCUUUUCCACUUCUGC___GCCUUCAUGCAAA ...(.(((((.((((.(....)))))..((...((((((......))))))...))(((....)))........................................))))))... (-18.42 = -19.02 + 0.60)

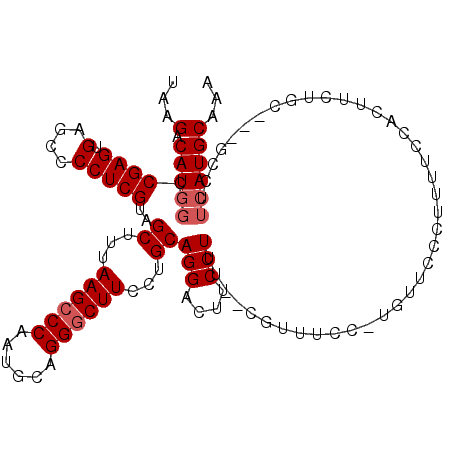
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:45:49 2006