
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,667,313 – 15,667,454 |
| Length | 141 |
| Max. P | 0.999532 |

| Location | 15,667,313 – 15,667,416 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -23.84 |
| Energy contribution | -23.70 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15667313 103 - 23771897 AUGGCUCAUUAGGGUCUUAAAUUAAAGCUACUUGGCCCAAGUCGGUCAACUAAGCCGACGUGCGGAGAAA-----------------GAAAAAGAAAGCGGCGCGAAGACAAGACCACAA ............((((((...((...((..(((..((((.((((((.......)))))).)).)).....-----------------........)))..))...))...)))))).... ( -25.16) >DroSec_CAF1 27019 103 - 1 AUGGCUCAUUAGGGUCUUAAAUUAAAGCUACUUGGCCCAAGUCGGUCAACUAAGCCGACGUGCGGAGAAA-----------------GAAAAAGAAAGCGGCGCGAAGACAAGACCACAA ............((((((...((...((..(((..((((.((((((.......)))))).)).)).....-----------------........)))..))...))...)))))).... ( -25.16) >DroSim_CAF1 27000 103 - 1 AUGGCUCAUUAGGGUCUUAAAUUAAAGCUACUUGGCCCAAGUCGGUCAACUAAGCCGACGUGCGGAGAAA-----------------GAAAAAGAAAGCGGCGCGAAGACAAGACCACAA ............((((((...((...((..(((..((((.((((((.......)))))).)).)).....-----------------........)))..))...))...)))))).... ( -25.16) >DroEre_CAF1 26594 102 - 1 AUGGCUCAUUAGGGUCUUAAAUUAAAGCUACUUGGC-CAAGUCGGUCAACUAAGCCGACGUGCGGAGAAA-----------------GAAAAAGAAAGCGGCGCGAAGACAAGACCACAA ............((((((...((...(((.(((.((-...((((((.......))))))..)).)))...-----------------......(....))))...))...)))))).... ( -25.00) >DroYak_CAF1 27193 103 - 1 AUGGCUCAUUAGGGUCUUAAAUUAAAGCUACUUGGCCCAAGUCGGUCAACUAAGCCGACGUGCGGAGAAA-----------------GAAAAAGAAAGCGGCGCGAAGACAAGACCACAA ............((((((...((...((..(((..((((.((((((.......)))))).)).)).....-----------------........)))..))...))...)))))).... ( -25.16) >DroAna_CAF1 85641 120 - 1 AUGGCUCAUUAGGGUCUUAAAUUAAAGCUACUUGGCCCAAGUCGGUCAACUAAGCCGACGUGCAAAGAAAUGGGAAAAUGGUAAAUGGGAAAAGAAAGCGGCCCGAAGACAAGACCACAA .(((((..((.((((((((...))).(((.(((..((((.((((((.......))))))...(..(....)..)...........))))..)))..))))))))...))..)).)))... ( -28.20) >consensus AUGGCUCAUUAGGGUCUUAAAUUAAAGCUACUUGGCCCAAGUCGGUCAACUAAGCCGACGUGCGGAGAAA_________________GAAAAAGAAAGCGGCGCGAAGACAAGACCACAA ............((((((...((...(((.(((.((....((((((.......))))))..)).)))..........................(....))))...))...)))))).... (-23.84 = -23.70 + -0.14)

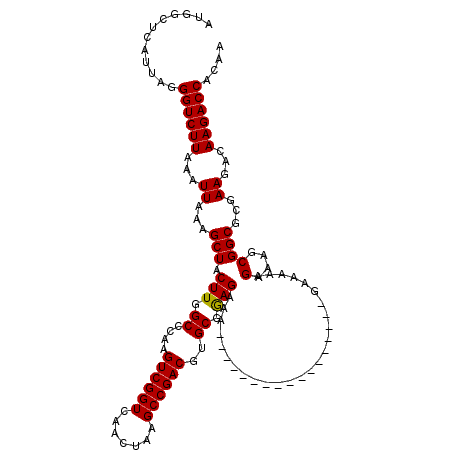

| Location | 15,667,346 – 15,667,454 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.87 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15667346 108 + 23771897 ----------UUUCUCCGCACGUCGGCUUAGUUGACCGACUUGGGCCAAGUAGCUUUAAUUUAAGACCCUAAUGAGCCAUUUAGGCAUUCUGGGCUUUGUGUCUUCAAUGCCGACUAC ----------...........((((((...(((((..(((..(((((.((...(((......)))...)).....(((.....)))......)))))...))).)))))))))))... ( -29.30) >DroSec_CAF1 27052 108 + 1 ----------UUUCUCCGCACGUCGGCUUAGUUGACCGACUUGGGCCAAGUAGCUUUAAUUUAAGACCCUAAUGAGCCAUUUAGGCAUUCUGGGCUUUGUGUCUUCAAUGCCGACUAC ----------...........((((((...(((((..(((..(((((.((...(((......)))...)).....(((.....)))......)))))...))).)))))))))))... ( -29.30) >DroSim_CAF1 27033 108 + 1 ----------UUUCUCCGCACGUCGGCUUAGUUGACCGACUUGGGCCAAGUAGCUUUAAUUUAAGACCCUAAUGAGCCAUUUAGGCAUUCUGGGCUUUGUGUCUUCAAUGCCGACUAC ----------...........((((((...(((((..(((..(((((.((...(((......)))...)).....(((.....)))......)))))...))).)))))))))))... ( -29.30) >DroEre_CAF1 26627 107 + 1 ----------UUUCUCCGCACGUCGGCUUAGUUGACCGACUUG-GCCAAGUAGCUUUAAUUUAAGACCCUAAUGAGCCAUUUAGGCAUUCUGGGCUUUGUGUCUUCAAUGCCGACUAC ----------...........(((((((.((((....)))).)-)))((((.((((....(((......))).)))).)))).((((((..((((.....))))..)))))))))... ( -29.80) >DroYak_CAF1 27226 108 + 1 ----------UUUCUCCGCACGUCGGCUUAGUUGACCGACUUGGGCCAAGUAGCUUUAAUUUAAGACCCUAAUGAGCCAUUUAGGCAUUCUGGGCUUUGUGUCUUCAAUGCCGACUAC ----------...........((((((...(((((..(((..(((((.((...(((......)))...)).....(((.....)))......)))))...))).)))))))))))... ( -29.30) >DroAna_CAF1 85681 118 + 1 CAUUUUCCCAUUUCUUUGCACGUCGGCUUAGUUGACCGACUUGGGCCAAGUAGCUUUAAUUUAAGACCCUAAUGAGCCAUUUAGGCAUUCUGGGCUUUGUGUCUUCAAUGCCGACUAC .....................((((((...(((((..(((..(((((.((...(((......)))...)).....(((.....)))......)))))...))).)))))))))))... ( -29.30) >consensus __________UUUCUCCGCACGUCGGCUUAGUUGACCGACUUGGGCCAAGUAGCUUUAAUUUAAGACCCUAAUGAGCCAUUUAGGCAUUCUGGGCUUUGUGUCUUCAAUGCCGACUAC .....................((((((...(((((..(((..(((((.((...(((......)))...)).....(((.....)))......)))))...))).)))))))))))... (-28.70 = -28.87 + 0.17)

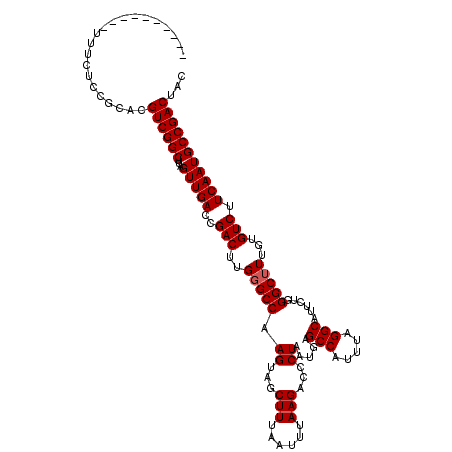

| Location | 15,667,346 – 15,667,454 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -31.27 |
| Energy contribution | -31.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15667346 108 - 23771897 GUAGUCGGCAUUGAAGACACAAAGCCCAGAAUGCCUAAAUGGCUCAUUAGGGUCUUAAAUUAAAGCUACUUGGCCCAAGUCGGUCAACUAAGCCGACGUGCGGAGAAA---------- (((((((((.((((.(((...((((((.((..(((.....)))...)).))).)))........(((....)))....)))..))))....)))))).))).......---------- ( -31.40) >DroSec_CAF1 27052 108 - 1 GUAGUCGGCAUUGAAGACACAAAGCCCAGAAUGCCUAAAUGGCUCAUUAGGGUCUUAAAUUAAAGCUACUUGGCCCAAGUCGGUCAACUAAGCCGACGUGCGGAGAAA---------- (((((((((.((((.(((...((((((.((..(((.....)))...)).))).)))........(((....)))....)))..))))....)))))).))).......---------- ( -31.40) >DroSim_CAF1 27033 108 - 1 GUAGUCGGCAUUGAAGACACAAAGCCCAGAAUGCCUAAAUGGCUCAUUAGGGUCUUAAAUUAAAGCUACUUGGCCCAAGUCGGUCAACUAAGCCGACGUGCGGAGAAA---------- (((((((((.((((.(((...((((((.((..(((.....)))...)).))).)))........(((....)))....)))..))))....)))))).))).......---------- ( -31.40) >DroEre_CAF1 26627 107 - 1 GUAGUCGGCAUUGAAGACACAAAGCCCAGAAUGCCUAAAUGGCUCAUUAGGGUCUUAAAUUAAAGCUACUUGGC-CAAGUCGGUCAACUAAGCCGACGUGCGGAGAAA---------- (((((((((............((((((.((..(((.....)))...)).))).))).............(((((-(.....))))))....)))))).))).......---------- ( -31.50) >DroYak_CAF1 27226 108 - 1 GUAGUCGGCAUUGAAGACACAAAGCCCAGAAUGCCUAAAUGGCUCAUUAGGGUCUUAAAUUAAAGCUACUUGGCCCAAGUCGGUCAACUAAGCCGACGUGCGGAGAAA---------- (((((((((.((((.(((...((((((.((..(((.....)))...)).))).)))........(((....)))....)))..))))....)))))).))).......---------- ( -31.40) >DroAna_CAF1 85681 118 - 1 GUAGUCGGCAUUGAAGACACAAAGCCCAGAAUGCCUAAAUGGCUCAUUAGGGUCUUAAAUUAAAGCUACUUGGCCCAAGUCGGUCAACUAAGCCGACGUGCAAAGAAAUGGGAAAAUG (((((((((.((((.(((...((((((.((..(((.....)))...)).))).)))........(((....)))....)))..))))....)))))).)))................. ( -31.40) >consensus GUAGUCGGCAUUGAAGACACAAAGCCCAGAAUGCCUAAAUGGCUCAUUAGGGUCUUAAAUUAAAGCUACUUGGCCCAAGUCGGUCAACUAAGCCGACGUGCGGAGAAA__________ (((((((((.((((.(((...((((((.((..(((.....)))...)).))).)))........(((....)))....)))..))))....)))))).)))................. (-31.27 = -31.27 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:55 2006