
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,661,721 – 15,661,927 |
| Length | 206 |
| Max. P | 0.952944 |

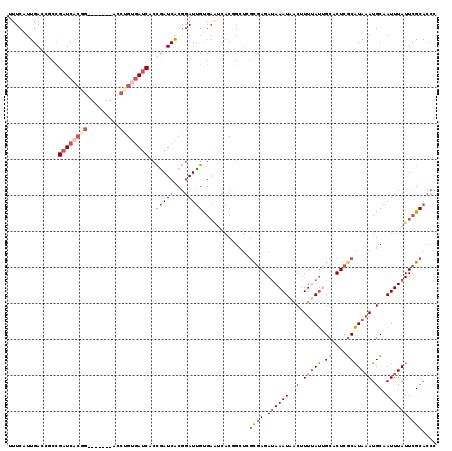
| Location | 15,661,721 – 15,661,833 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.43 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -12.95 |
| Energy contribution | -14.23 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15661721 112 + 23771897 UUUCAUUGACCGCUGAUCACGG-------ACCUGUGAUCACCAAUCACGGAUUGUGAAUCACGGCUUGCGAGAUAAAUAACUUUUAUUGCACUCGCAUAAAUGCAAUUUAUUCGCACCC .........(((.(((((((((-------.((.(((((.....))))))).))))).)))))))..((((((.(((((..(.(((((.((....))))))).)..)))))))))))... ( -30.30) >DroSec_CAF1 21533 112 + 1 UUUCAUUGAGCGCCGAUCACGG-------ACCUGUGAUCACCGAUCACGGAUUGUGAAUCACGGCUAGCGAGAUAAAUAACUUUUAUUGCACUCGCAUAAAUGCAAUUUAUUCGCACCC ...........((((.((((((-------.((.((((((...)))))))).))))))....))))..(((((.(((((..(.(((((.((....))))))).)..)))))))))).... ( -31.80) >DroSim_CAF1 21504 112 + 1 UUUCAUUGACCGCCGAUCACGG-------ACCUGUGAUCACCGAUCACGGAUUGUGAAUCACGGCUAGCGAGAUAAAUAACUUUUAUUGCACUCGCAUAAAUGCAAUUUAUUCGCACCC ...........((((.((((((-------.((.((((((...)))))))).))))))....))))..(((((.(((((..(.(((((.((....))))))).)..)))))))))).... ( -31.80) >DroEre_CAF1 21359 112 + 1 UUUCAUUGAUCACCGCUCACCG-------AUCGGCGAUCACUGAUCACGGAUUGCGAGUCACGGCUCGUGAGAAAAAUAACUUUUAUUGCACUCGCAUAAAUGCAAUUUAUUCGCACCC ..(((.(((((.(((.((...)-------).))).))))).)))....(((((((((((....))))(((((...((((.....))))...)))))......))))))).......... ( -31.60) >DroYak_CAF1 21568 119 + 1 UUUCAUUGAUCACCGAUCACUGGUCACGGACCUGUGAUCACCGAUCACGGAUUGCGAGGCACUGGUCGUGAGAUAAAUAACUUUAAUGGCACUCGCAUAAAUGCAAUUUAUUCGCACCC ......(((((...((((((.((((...)))).))))))...))))).((..((((((((....(((....)))......(......))).))))))....(((.........))))). ( -35.60) >DroAna_CAF1 80448 94 + 1 UUUCAAUGAACCACGAUCCCCG--------------AUCCCCUAUCGCCCAUAGU-----------CAUAAGAUAAAUUGCCUUUACGGCGCUCCCGUAUAUGCGAUUUAUUCGCAUUA .....((((.....((((...)--------------)))..((((.....)))))-----------)))..((((((((((...(((((.....)))))...))))))))))....... ( -18.10) >consensus UUUCAUUGACCGCCGAUCACGG_______ACCUGUGAUCACCGAUCACGGAUUGUGAAUCACGGCUCGCGAGAUAAAUAACUUUUAUUGCACUCGCAUAAAUGCAAUUUAUUCGCACCC ..............((((((((.........))))))))..(((((...))))).............((((.((((((..(.(((((.((....))))))).)..)))))))))).... (-12.95 = -14.23 + 1.28)


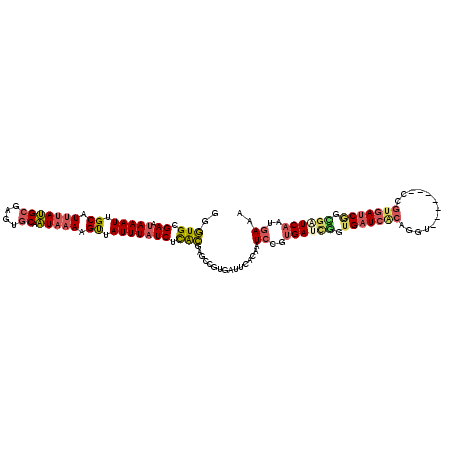
| Location | 15,661,721 – 15,661,833 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.43 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -20.88 |
| Energy contribution | -21.59 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15661721 112 - 23771897 GGGUGCGAAUAAAUUGCAUUUAUGCGAGUGCAAUAAAAGUUAUUUAUCUCGCAAGCCGUGAUUCACAAUCCGUGAUUGGUGAUCACAGGU-------CCGUGAUCAGCGGUCAAUGAAA ((.(((((((((((.((.(((((((....)).))))).)).)))))).)))))..))(((...)))..((..((((((.(((((((....-------..))))))).))))))..)).. ( -36.20) >DroSec_CAF1 21533 112 - 1 GGGUGCGAAUAAAUUGCAUUUAUGCGAGUGCAAUAAAAGUUAUUUAUCUCGCUAGCCGUGAUUCACAAUCCGUGAUCGGUGAUCACAGGU-------CCGUGAUCGGCGCUCAAUGAAA ((((((((((((((.((.(((((((....)).))))).)).)))))).))))..((((....((((...((((((((...)))))).)).-------..)))).))))))))....... ( -36.80) >DroSim_CAF1 21504 112 - 1 GGGUGCGAAUAAAUUGCAUUUAUGCGAGUGCAAUAAAAGUUAUUUAUCUCGCUAGCCGUGAUUCACAAUCCGUGAUCGGUGAUCACAGGU-------CCGUGAUCGGCGGUCAAUGAAA ((((((((((((((.((.(((((((....)).))))).)).)))))).)))).....(((...))).)))).((((((.(((((((....-------..))))))).))))))...... ( -36.30) >DroEre_CAF1 21359 112 - 1 GGGUGCGAAUAAAUUGCAUUUAUGCGAGUGCAAUAAAAGUUAUUUUUCUCACGAGCCGUGACUCGCAAUCCGUGAUCAGUGAUCGCCGAU-------CGGUGAGCGGUGAUCAAUGAAA ((((((((....(((((((((....)))))))))....(((((..(((....)))..))))))))).)))).((((((.((.(((((...-------.))))).)).))))))...... ( -39.20) >DroYak_CAF1 21568 119 - 1 GGGUGCGAAUAAAUUGCAUUUAUGCGAGUGCCAUUAAAGUUAUUUAUCUCACGACCAGUGCCUCGCAAUCCGUGAUCGGUGAUCACAGGUCCGUGACCAGUGAUCGGUGAUCAAUGAAA ((((((((.....)))).....((((((.((...((((....))))....((.....)))))))))))))).((((((.(((((((.((((...)))).))))))).))))))...... ( -39.60) >DroAna_CAF1 80448 94 - 1 UAAUGCGAAUAAAUCGCAUAUACGGGAGCGCCGUAAAGGCAAUUUAUCUUAUG-----------ACUAUGGGCGAUAGGGGAU--------------CGGGGAUCGUGGUUCAUUGAAA ..((((((.....)))))).(((((.....)))))...........((..(((-----------(((((((.((((.....))--------------))....)))))).)))).)).. ( -22.60) >consensus GGGUGCGAAUAAAUUGCAUUUAUGCGAGUGCAAUAAAAGUUAUUUAUCUCACGAGCCGUGAUUCACAAUCCGUGAUCGGUGAUCACAGGU_______CCGUGAUCGGCGAUCAAUGAAA ..(((.((.(((((.((.(((((((....)).))))).)).))))))).)))................((..((((((.(((((((.............))))))).))))))..)).. (-20.88 = -21.59 + 0.70)

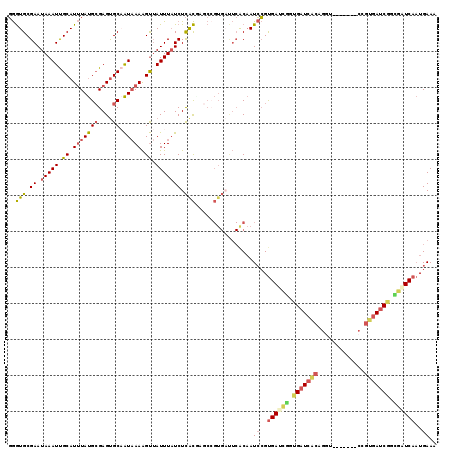
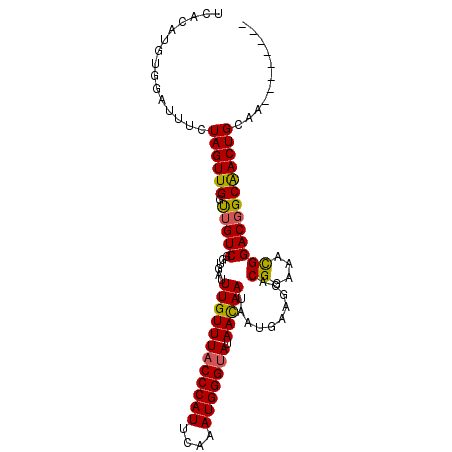
| Location | 15,661,833 – 15,661,927 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 92.59 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -23.54 |
| Energy contribution | -23.14 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15661833 94 + 23771897 UCACAUGUGGAUUUCUAGUUGGUUGUCGGUGAUUUGUUUACCCAUUCAAAUGGGUAUAACAAUAAUGAAGCACGGAAACGGACGGCGACUGCAA-------- ...((..(((....)))..))(((((((.....((((((((((((....))))))).)))))..........((....))..))))))).....-------- ( -26.70) >DroSec_CAF1 21645 94 + 1 UCACAUGUGGAUUUCUAGUUGGUUGUCGGUGAUUUGUUUACCCAUUCAAAUGGGUAUAACAAUAAUGAAGCACGGAAACGGACGGCAACUGCAA-------- ...((..(((....)))..))(((((((.....((((((((((((....))))))).)))))..........((....))..))))))).....-------- ( -27.00) >DroSim_CAF1 21616 94 + 1 UCACAUGUGGAUUUCUAGUUGGUUGUCGGUGAUUUGUUUACCCAUUCAAAUGGGUAUAACAAUAAUGAAGCACGGAAACGGACGGCGACUGCAA-------- ...((..(((....)))..))(((((((.....((((((((((((....))))))).)))))..........((....))..))))))).....-------- ( -26.70) >DroEre_CAF1 21471 94 + 1 UCACAUGUGGCUUUCUAGUUGGCUGUCGGUGAUUUGUUUACCCAUUAAAAUGGGCAUAAUAAUAAUGAAGCACGGAAAUGGACGGCAACUGCAA-------- ...............((((((.(((((.((..(.(((((.(((((....)))))(((.......)))))))).)...)).)))))))))))...-------- ( -21.90) >DroYak_CAF1 21687 102 + 1 UCACAUGUGGAUUUCUAGUUGGUUGUCGGUGAUUUGUUUACCCAUUCAAAUGGGUAUAAUAAUAAUGAAGCACGGAAGCGGACUGCAACUGCAAGUGCAUUG .(((.((..(.....(((((.(((.((.(((.((..(((((((((....))))))).......))..)).))).))))).)))))...)..)).)))..... ( -27.71) >consensus UCACAUGUGGAUUUCUAGUUGGUUGUCGGUGAUUUGUUUACCCAUUCAAAUGGGUAUAACAAUAAUGAAGCACGGAAACGGACGGCAACUGCAA________ ...............((((((.(((((......((((((((((((....))))))).)))))..........((....)))))))))))))........... (-23.54 = -23.14 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:44 2006