
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,649,504 – 15,649,611 |
| Length | 107 |
| Max. P | 0.802188 |

| Location | 15,649,504 – 15,649,611 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.43 |
| Mean single sequence MFE | -38.83 |
| Consensus MFE | -24.09 |
| Energy contribution | -25.52 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
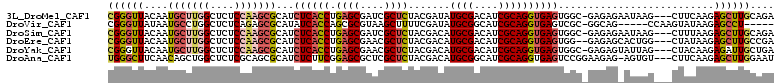
>3L_DroMel_CAF1 15649504 107 + 23771897 CGGGUUACAAUGCUUGGCUCUCCAAGCGCAUCUCACCUGAGCGAUCGCUCUACGAUAUGCGACAUCGCAGGUGAGUGGC-GAGAGAAUAAG---CUUCAAGAGCUUGCAGA ((((((.....(((((..((((....(((..(((((((((((....))))........(((....))))))))))..))-).)))).))))---)......)))))).... ( -40.00) >DroVir_CAF1 71786 100 + 1 CGGGUUAUAAUGCCUGGCUCUCAGAGCGCAUAUCACCAGCGCGUAAGCUUUUCGAUAUGCGGCAUCGCAGGUGAGUCGC-GGCAG-----CCAAGUAUAAGAGCCU----- .(((((...((((.(((((((....(((....(((((...((....)).........((((....)))))))))..)))-)).))-----))).))))...)))))----- ( -35.10) >DroSim_CAF1 9196 107 + 1 CGGGUUACAAUGCUUGGCUCUCCAAGCGCAUCUCACCUGAGCGAUCGCUCUACGACAUGCGACAUCGCAGGUGAGUGGC-GAGAGAAUAAG---CUUUAAGAGCUUGCAGA ((((((.....(((((..((((....(((..(((((((((((....))))........(((....))))))))))..))-).)))).))))---)......)))))).... ( -40.00) >DroEre_CAF1 9624 106 + 1 CGGGUUACAAUGCUUGGCUCUCCAAGCGCAUCUCACCUGAGCGAACGCUCUACGACAUGCGACAUCGCAGGUGAGUGG--GAGAGCACUGG---CUAUAAGAGCUUGCCGA ((((((.....(((..(((((((.....((.(((((((((((....))))........(((....)))))))))))))--))))))...))---)......)))))).... ( -41.60) >DroYak_CAF1 9255 107 + 1 CGGGUUACAAUGCUUGGCUCUCCAAGCGCAUCUCACCUGAGCGAACGCUCUACGACAUGCGACAUCGCAGGUGAGUGGC-GAGAGUAUUAG---CUACAAGAGAUUGCUGA (((((......)))))((((((...((.((.(((((((((((....))))........(((....))))))))))))))-)))))).((((---(...........))))) ( -38.10) >DroAna_CAF1 68847 107 + 1 UGGGCUUCAACAGCUGGCUCUCGCAGCGCAUCUCUUCGGAGCGCUCGCUCUACGACAUGCGGCAUCGCAGGUGAGUCCGGAAGAG-AGUGU---CUUCAAGAGCUUGGAAU ..((((.....))))((((((.(.(((((.((((((((((((....)))))..(((.((((....)))).....)))..))))))-)))).---)).).))))))...... ( -38.20) >consensus CGGGUUACAAUGCUUGGCUCUCCAAGCGCAUCUCACCUGAGCGAACGCUCUACGACAUGCGACAUCGCAGGUGAGUGGC_GAGAG_AUUAG___CUACAAGAGCUUGCAGA ((((((....(((((((....)))))))...((((((.((((....)))).......((((....))))))))))..........................)))))).... (-24.09 = -25.52 + 1.42)
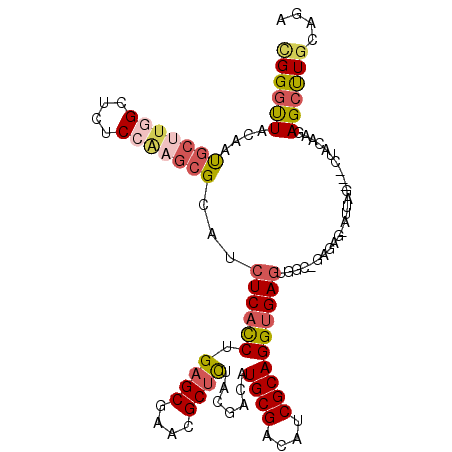
| Location | 15,649,504 – 15,649,611 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.43 |
| Mean single sequence MFE | -38.17 |
| Consensus MFE | -22.65 |
| Energy contribution | -24.32 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
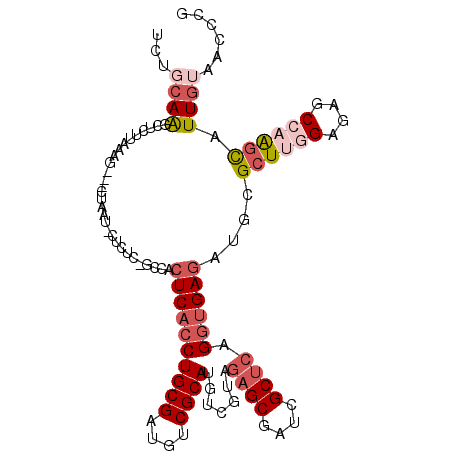
>3L_DroMel_CAF1 15649504 107 - 23771897 UCUGCAAGCUCUUGAAG---CUUAUUCUCUC-GCCACUCACCUGCGAUGUCGCAUAUCGUAGAGCGAUCGCUCAGGUGAGAUGCGCUUGGAGAGCCAAGCAUUGUAACCCG ..(((..((((((.(((---(.((...((((-(((.......((((((((...))))))))((((....)))).))))))))).)))).))))))...))).......... ( -40.80) >DroVir_CAF1 71786 100 - 1 -----AGGCUCUUAUACUUGG-----CUGCC-GCGACUCACCUGCGAUGCCGCAUAUCGAAAAGCUUACGCGCUGGUGAUAUGCGCUCUGAGAGCCAGGCAUUAUAACCCG -----.((((((((.....((-----(...(-(((.......))))..)))((((((((...(((......)))..))))))))....))))))))............... ( -36.50) >DroSim_CAF1 9196 107 - 1 UCUGCAAGCUCUUAAAG---CUUAUUCUCUC-GCCACUCACCUGCGAUGUCGCAUGUCGUAGAGCGAUCGCUCAGGUGAGAUGCGCUUGGAGAGCCAAGCAUUGUAACCCG ...(((((((.....))---)))....((((-(((((..((.((((....)))).)).)).((((....)))).))))))).))((((((....))))))........... ( -38.60) >DroEre_CAF1 9624 106 - 1 UCGGCAAGCUCUUAUAG---CCAGUGCUCUC--CCACUCACCUGCGAUGUCGCAUGUCGUAGAGCGUUCGCUCAGGUGAGAUGCGCUUGGAGAGCCAAGCAUUGUAACCCG ..(((...........)---)).(.((((((--(((((((((((((....)))........((((....))))))))))).)).....))))))))............... ( -37.50) >DroYak_CAF1 9255 107 - 1 UCAGCAAUCUCUUGUAG---CUAAUACUCUC-GCCACUCACCUGCGAUGUCGCAUGUCGUAGAGCGUUCGCUCAGGUGAGAUGCGCUUGGAGAGCCAAGCAUUGUAACCCG ...(((.......(((.---....)))((((-(((((..((.((((....)))).)).)).((((....)))).))))))))))((((((....))))))........... ( -36.40) >DroAna_CAF1 68847 107 - 1 AUUCCAAGCUCUUGAAG---ACACU-CUCUUCCGGACUCACCUGCGAUGCCGCAUGUCGUAGAGCGAGCGCUCCGAAGAGAUGCGCUGCGAGAGCCAGCUGUUGAAGCCCA .......(((((((.((---.(.((-((((((.((((((.((((((((.......))))))).).)))...)))))))))).).))).)))))))..(((.....)))... ( -39.20) >consensus UCUGCAAGCUCUUAAAG___CUAAU_CUCUC_GCCACUCACCUGCGAUGUCGCAUGUCGUAGAGCGAUCGCUCAGGUGAGAUGCGCUUGGAGAGCCAAGCAUUGUAACCCG ...((((.............................((((((((((....)))).......((((....)))).))))))....((((((....)))))).))))...... (-22.65 = -24.32 + 1.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:32 2006