
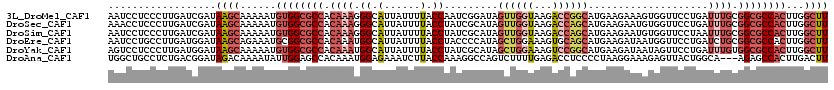
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,647,270 – 15,647,390 |
| Length | 120 |
| Max. P | 0.963267 |

| Location | 15,647,270 – 15,647,390 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.50 |
| Mean single sequence MFE | -36.97 |
| Consensus MFE | -19.05 |
| Energy contribution | -21.00 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15647270 120 + 23771897 AAUCCUCCCUUGAUCGAUAAGCAAAAAUGUGGCGCCACAAAGGGCAUUAUUUUACCAAUCGGAUAGUUGGUAAGACCGGCAUGAAGAAAGUGGUUCCUGAUUUGCGGCGCCACUUGGCUU ..................((((...((.((((((((.(((((((.((((((((.((....))...(((((.....)))))......)))))))).)))..)))).)))))))))).)))) ( -38.40) >DroSec_CAF1 6918 120 + 1 AAACCUCCCUUGAUCGAUAAGCAAAAAUGUGGCGCCACAAAGGGCAUUAUUUUACCUAUCGCAUAGUUGGUAAGACCAGCAUGAAGAAUGUGGUUCCUGAUUUGCGGCGCCACUUGGCUU ..................((((...((.((((((((....(((...........)))...(((.(((..(...(((((.(((.....)))))))).)..)))))))))))))))).)))) ( -36.30) >DroSim_CAF1 6931 120 + 1 AAUCCUCCCUUGAUCGAUAAGCAAAAAUGUGGCGCCACAAAGGGCAUUAUUUUACCUAUCGCAUAGUUGGUAAGACCAGCAUGAAGAAUGUGGUUCCUAAUUUGCGGCGCCACUUGGCUU ..................((((...((.((((((((....(((...........)))...(((.((((((...(((((.(((.....)))))))).))))))))))))))))))).)))) ( -36.80) >DroEre_CAF1 7339 120 + 1 AAUCCUGCCUUGAUGGAUAAGCAGAAAUGCGGCGCCACAAAUGGCAUUAUUUUACCUACCCCAUAGCUGGAAAGUGCAGCAUGAAGAUAAUGGUUCCUGAUCUGCGGCGCCACUUGGCUU ......(((..(........(((....)))((((((.((...(((((((((((........(((.((((.......))))))))))))))))...)).....)).)))))).)..))).. ( -36.00) >DroYak_CAF1 6975 120 + 1 AGUCCUCCCUUGAUGGAUAAGCAAAAAUGUGGCGCCACAAAUGGCAUUAUUUUACCUAUCGCAUAGCUGGAAAGUCCGGCAUGAAGAUAAUAGUUCCUGAUUUGUGGCGCCACUUGGCUU .((((((....)).))))((((...((.((((((((((((((((.(((((......((((.(((.(((((.....))))))))..))))))))).))..)))))))))))))))).)))) ( -46.20) >DroAna_CAF1 66686 117 + 1 UGGCUGCCUCUGACGGAUAGACAAAAUAUUGGAGCCACAAAUGGAGAAAUCUUACCAAAGGCCAGUCUUUUGAGACCUCCCCUAAGGAAAGAGUUACUGGCA---AGAGCCACUUGACUU ((((((((..((((.......(((((.(((((..........(((....))).........))))).)))))...(((......))).....))))..))).---..)))))........ ( -28.11) >consensus AAUCCUCCCUUGAUCGAUAAGCAAAAAUGUGGCGCCACAAAGGGCAUUAUUUUACCUAUCGCAUAGCUGGUAAGACCAGCAUGAAGAAAAUGGUUCCUGAUUUGCGGCGCCACUUGGCUU ..................((((......((((((((.((((.((.(......).)).........((((((...))))))....................)))).))))))))...)))) (-19.05 = -21.00 + 1.95)


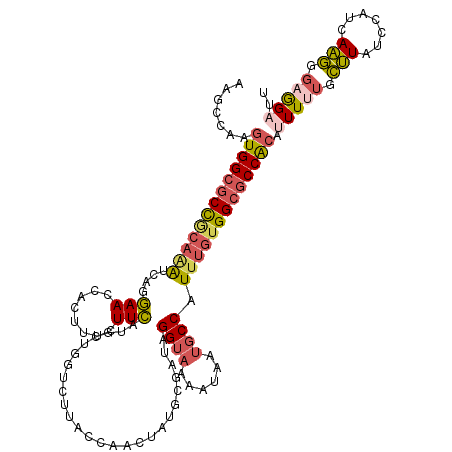
| Location | 15,647,270 – 15,647,390 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.50 |
| Mean single sequence MFE | -39.17 |
| Consensus MFE | -23.57 |
| Energy contribution | -24.30 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15647270 120 - 23771897 AAGCCAAGUGGCGCCGCAAAUCAGGAACCACUUUCUUCAUGCCGGUCUUACCAACUAUCCGAUUGGUAAAAUAAUGCCCUUUGUGGCGCCACAUUUUUGCUUAUCGAUCAAGGGAGGAUU .......(((((((((((((..(((((.....)))))..((((((((.............))))))))...........)))))))))))))((((((.(((.......))).)))))). ( -36.72) >DroSec_CAF1 6918 120 - 1 AAGCCAAGUGGCGCCGCAAAUCAGGAACCACAUUCUUCAUGCUGGUCUUACCAACUAUGCGAUAGGUAAAAUAAUGCCCUUUGUGGCGCCACAUUUUUGCUUAUCGAUCAAGGGAGGUUU (((((..(((((((((((((..((((.((((((.....))).)))))))...............((((......)))).)))))))))))))....((.(((.......))).))))))) ( -37.90) >DroSim_CAF1 6931 120 - 1 AAGCCAAGUGGCGCCGCAAAUUAGGAACCACAUUCUUCAUGCUGGUCUUACCAACUAUGCGAUAGGUAAAAUAAUGCCCUUUGUGGCGCCACAUUUUUGCUUAUCGAUCAAGGGAGGAUU .......(((((((((((((.(((((.((((((.....))).))))))))..............((((......)))).)))))))))))))((((((.(((.......))).)))))). ( -37.70) >DroEre_CAF1 7339 120 - 1 AAGCCAAGUGGCGCCGCAGAUCAGGAACCAUUAUCUUCAUGCUGCACUUUCCAGCUAUGGGGUAGGUAAAAUAAUGCCAUUUGUGGCGCCGCAUUUCUGCUUAUCCAUCAAGGCAGGAUU .......((((((((((((((.............((((((((((.......)))).))))))..((((......))))))))))))))))))..((((((((........)))))))).. ( -46.50) >DroYak_CAF1 6975 120 - 1 AAGCCAAGUGGCGCCACAAAUCAGGAACUAUUAUCUUCAUGCCGGACUUUCCAGCUAUGCGAUAGGUAAAAUAAUGCCAUUUGUGGCGCCACAUUUUUGCUUAUCCAUCAAGGGAGGACU .((((..((((((((((((((..........((((..(((((.((.....)).)).))).))))((((......))))))))))))))))))...........(((......))))).)) ( -42.00) >DroAna_CAF1 66686 117 - 1 AAGUCAAGUGGCUCU---UGCCAGUAACUCUUUCCUUAGGGGAGGUCUCAAAAGACUGGCCUUUGGUAAGAUUUCUCCAUUUGUGGCUCCAAUAUUUUGUCUAUCCGUCAGAGGCAGCCA ....(((((((.(((---(((((....((((......))))((((((..........)))))))))))))).....)))))))(((((((.....((((.(.....).)))))).))))) ( -34.20) >consensus AAGCCAAGUGGCGCCGCAAAUCAGGAACCACUUUCUUCAUGCUGGUCUUACCAACUAUGCGAUAGGUAAAAUAAUGCCAUUUGUGGCGCCACAUUUUUGCUUAUCCAUCAAGGGAGGAUU .......(((((((((((((....(((........)))..........................((((......)))).))))))))))))).(((((.(((.......))).))))).. (-23.57 = -24.30 + 0.73)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:30 2006