
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,636,188 – 15,636,338 |
| Length | 150 |
| Max. P | 0.982000 |

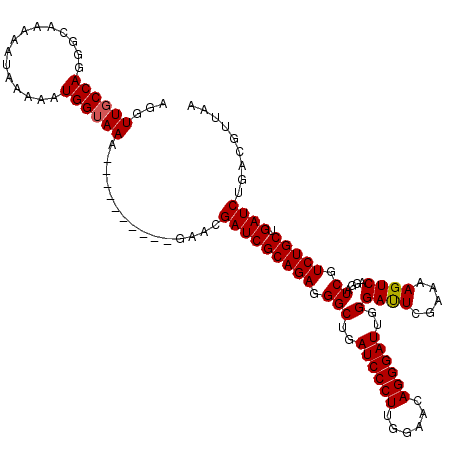
| Location | 15,636,188 – 15,636,298 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.03 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -31.20 |
| Energy contribution | -31.26 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15636188 110 + 23771897 AGGUUGCCAGGGCAAAAAUAAAAAUGGUAAA----------GAACGAUCGCAGAGGGCUGAUCCCUUGGAACAGGGAUUGGGAUUCGAAAAGUCAGGAUCGUCUGCUGAUCUGACGUUAA ...((((((...............)))))).----------....(((((((((...(..((((((......))))))..)(((((.........))))).))))).))))......... ( -32.06) >DroSec_CAF1 58295 110 + 1 AGGUUGCCAGGGCAAAAAUAAAAAUGGUAAA----------GAACGAUCGCAGAGGGCUGAUCCCUUGGAACAGGGAUUGGGAUUCGAAAAGUCAGGAUCGUCUGCUGAUCUGACGUUAA ...((((((...............)))))).----------....(((((((((...(..((((((......))))))..)(((((.........))))).))))).))))......... ( -32.06) >DroSim_CAF1 59263 110 + 1 AGGUUGCCAGGGCAAAAAUAAAAAUGGUAAA----------GAACGAUCGCAGAGGGCUGAUCCCUUGGAACAGGGAUUGGGAUUCGAGAAGUCAGGAUCGUCUGCUGAUCUGACGUUAA ...((((((...............)))))).----------....(((((((((...(..((((((......))))))..)(((((.........))))).))))).))))......... ( -32.06) >DroYak_CAF1 63205 120 + 1 AGGUUGCCAGGGCCAAAGUAGAAAUGGAAAAAGGAGGAAUGGAACGAUCGCAGAGGGCUGAUCCCUCGGAACAGGGAUUGGGACUGGAGAAGUCAGGAUCGUCUGCUGAUCUGACGUUAA ..(((.(((...((.....................))..))))))(((((((((.(((..((((((......))))))..)((((.....))))....)).))))).))))......... ( -35.80) >consensus AGGUUGCCAGGGCAAAAAUAAAAAUGGUAAA__________GAACGAUCGCAGAGGGCUGAUCCCUUGGAACAGGGAUUGGGAUUCGAAAAGUCAGGAUCGUCUGCUGAUCUGACGUUAA ...((((((...............))))))...............(((((((((.(((..((((((......))))))..)((((.....))))....)).))))).))))......... (-31.20 = -31.26 + 0.06)

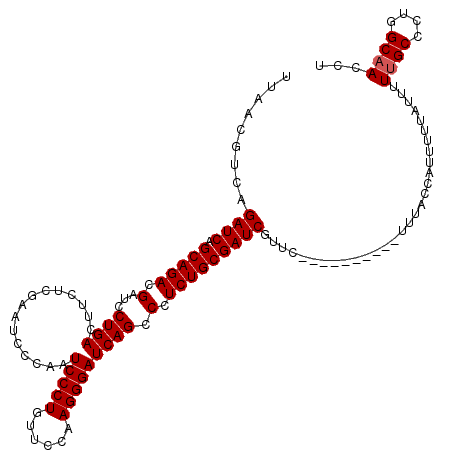
| Location | 15,636,188 – 15,636,298 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.03 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -22.68 |
| Energy contribution | -22.93 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15636188 110 - 23771897 UUAACGUCAGAUCAGCAGACGAUCCUGACUUUUCGAAUCCCAAUCCCUGUUCCAAGGGAUCAGCCCUCUGCGAUCGUUC----------UUUACCAUUUUUAUUUUUGCCCUGGCAACCU .........((((.((((((((..........))).......((((((......))))))......)))))))))....----------................((((....))))... ( -24.90) >DroSec_CAF1 58295 110 - 1 UUAACGUCAGAUCAGCAGACGAUCCUGACUUUUCGAAUCCCAAUCCCUGUUCCAAGGGAUCAGCCCUCUGCGAUCGUUC----------UUUACCAUUUUUAUUUUUGCCCUGGCAACCU .........((((.((((((((..........))).......((((((......))))))......)))))))))....----------................((((....))))... ( -24.90) >DroSim_CAF1 59263 110 - 1 UUAACGUCAGAUCAGCAGACGAUCCUGACUUCUCGAAUCCCAAUCCCUGUUCCAAGGGAUCAGCCCUCUGCGAUCGUUC----------UUUACCAUUUUUAUUUUUGCCCUGGCAACCU .........((((.(((((.(...((((......(.....)..(((((......))))))))).).)))))))))....----------................((((....))))... ( -24.20) >DroYak_CAF1 63205 120 - 1 UUAACGUCAGAUCAGCAGACGAUCCUGACUUCUCCAGUCCCAAUCCCUGUUCCGAGGGAUCAGCCCUCUGCGAUCGUUCCAUUCCUCCUUUUUCCAUUUCUACUUUGGCCCUGGCAACCU .....(((((.......(((((((..((((.....))))..............(((((.....)))))...)))))))...............(((.........)))..)))))..... ( -26.10) >consensus UUAACGUCAGAUCAGCAGACGAUCCUGACUUCUCGAAUCCCAAUCCCUGUUCCAAGGGAUCAGCCCUCUGCGAUCGUUC__________UUUACCAUUUUUAUUUUUGCCCUGGCAACCU .........((((.(((((.(...((((...............(((((......))))))))).).)))))))))..............................((((....))))... (-22.68 = -22.93 + 0.25)


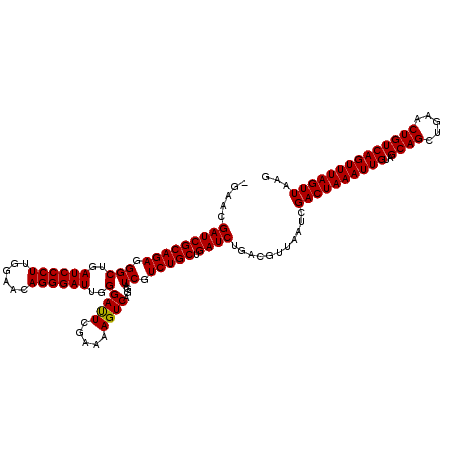
| Location | 15,636,219 – 15,636,338 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -40.47 |
| Consensus MFE | -40.06 |
| Energy contribution | -39.88 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15636219 119 + 23771897 -GAACGAUCGCAGAGGGCUGAUCCCUUGGAACAGGGAUUGGGAUUCGAAAAGUCAGGAUCGUCUGCUGAUCUGACGUUAAUCGACUAAAUUGUAGCAGCUGAACUGUCAGUUUAGUUAAG -....(((((((((...(..((((((......))))))..)(((((.........))))).))))).))))...........((((((((((..((((.....))))))))))))))... ( -39.80) >DroSec_CAF1 58326 119 + 1 -GAACGAUCGCAGAGGGCUGAUCCCUUGGAACAGGGAUUGGGAUUCGAAAAGUCAGGAUCGUCUGCUGAUCUGACGUUAAUCGACUAAAUUGUAGCAGCUGAACUGUCAGUUUAGUUAAG -....(((((((((...(..((((((......))))))..)(((((.........))))).))))).))))...........((((((((((..((((.....))))))))))))))... ( -39.80) >DroSim_CAF1 59294 119 + 1 -GAACGAUCGCAGAGGGCUGAUCCCUUGGAACAGGGAUUGGGAUUCGAGAAGUCAGGAUCGUCUGCUGAUCUGACGUUAAUCGACUAAAUUGUAGCAGCUGAACUGUCAGUUUAGUUAAG -....(((((((((...(..((((((......))))))..)(((((.........))))).))))).))))...........((((((((((..((((.....))))))))))))))... ( -39.80) >DroYak_CAF1 63245 120 + 1 GGAACGAUCGCAGAGGGCUGAUCCCUCGGAACAGGGAUUGGGACUGGAGAAGUCAGGAUCGUCUGCUGAUCUGACGUUAAUCGACUAAAUUGUAGCAGCUGAACUGUCAGUUUAGUUAAG .....(((((((((.(((..((((((......))))))..)((((.....))))....)).))))).))))...........((((((((((..((((.....))))))))))))))... ( -42.50) >consensus _GAACGAUCGCAGAGGGCUGAUCCCUUGGAACAGGGAUUGGGAUUCGAAAAGUCAGGAUCGUCUGCUGAUCUGACGUUAAUCGACUAAAUUGUAGCAGCUGAACUGUCAGUUUAGUUAAG .....(((((((((.(((..((((((......))))))..)((((.....))))....)).))))).))))...........((((((((((..((((.....))))))))))))))... (-40.06 = -39.88 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:22 2006