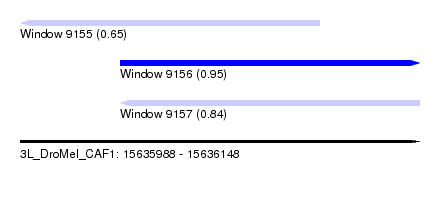
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,635,988 – 15,636,148 |
| Length | 160 |
| Max. P | 0.950883 |

| Location | 15,635,988 – 15,636,108 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -41.45 |
| Consensus MFE | -33.39 |
| Energy contribution | -34.45 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15635988 120 - 23771897 CAAAAUCACUAAGUGGCCCCGAGUGCCGCAGGUAGCUGCCAGACGGCAGGAGGCACGAUCAUUGAUCAAUGAUCAUCAGGACAAGGGGCAAUGCAGCACAGGUCGCGAUUGUACACUAAG ........((.(((((((((..(((((((.....))((((....))))...)))))(((((((....)))))))..........)))))..(((.((....)).)))......)))).)) ( -43.10) >DroSec_CAF1 58096 120 - 1 CAAAAUCACUAAGUGGACCCGAGUGCCGCAGGUAGCUGCCAGACGGCAGGAGGCACGAUCAUUGAUCAAUGAUCAUCAGGACAAGGGGCAAUACAGCACAGGUCGCGAUUGUACACUCAG ........((.((((..(((..(((((((.....))((((....))))...)))))(((((((....)))))))..........)))(((((...((.......)).))))).)))).)) ( -38.90) >DroSim_CAF1 59063 120 - 1 CAAAAUCACUAAGUGGCCCCGAGUGCCGCAGGUAGCUGCCAGACGGCAGGAGGCACGAUCAUUGUUCAAUGAUCAUCAGGACAAGGGGCACUCCAGCACAGGUCGCGAUUGUACACUCAG ........((.(((((((((..(((((((.....))((((....))))...)))))(((((((....)))))))..........)))))((....((.......))....)).)))).)) ( -41.70) >DroYak_CAF1 63009 116 - 1 CAAAAUCACUAAGUGGCCCUGAGUGCCGCAGGUAG---CCAGACGGCAGGAGGCACGAUCAUUGAUCAAUGAUCAUCGGGACAAGGG-CCAUCCAGCACAGGUCGCGAUUGUACACUUAG ........(((((((..((((((((((.(.....(---((....)))..).)))))(((((((....))))))).)))))((((..(-(...((......))..))..)))).))))))) ( -42.10) >consensus CAAAAUCACUAAGUGGCCCCGAGUGCCGCAGGUAGCUGCCAGACGGCAGGAGGCACGAUCAUUGAUCAAUGAUCAUCAGGACAAGGGGCAAUCCAGCACAGGUCGCGAUUGUACACUCAG ........((.(((((((((..(((((.(......(((((....)))))).)))))(((((((....)))))))..........)))))......((.......)).......)))).)) (-33.39 = -34.45 + 1.06)



| Location | 15,636,028 – 15,636,148 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -38.85 |
| Energy contribution | -39.35 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15636028 120 + 23771897 CCUGAUGAUCAUUGAUCAAUGAUCGUGCCUCCUGCCGUCUGGCAGCUACCUGCGGCACUCGGGGCCACUUAGUGAUUUUGGCCGUUGUUUGUUGACCUGGCCAAACAGCUGCCACUACUU (((((.(((((((....)))))))(((((...((((....))))((.....)))))))))))).......((((....((((.((((((((..........)))))))).)))).)))). ( -43.80) >DroSec_CAF1 58136 120 + 1 CCUGAUGAUCAUUGAUCAAUGAUCGUGCCUCCUGCCGUCUGGCAGCUACCUGCGGCACUCGGGUCCACUUAGUGAUUUUGGCCGUUGUUUGUUGACCAUGCCAAACAGCUGCCACUACUU (((((.(((((((....)))))))(((((...((((....))))((.....)))))))))))).......((((....((((.((((((((..........)))))))).)))).)))). ( -43.10) >DroSim_CAF1 59103 120 + 1 CCUGAUGAUCAUUGAACAAUGAUCGUGCCUCCUGCCGUCUGGCAGCUACCUGCGGCACUCGGGGCCACUUAGUGAUUUUGGCCGUUGUUUGUUGACCAGGCCAAACAGCUGCCACUACUU (((((.(((((((....)))))))(((((...((((....))))((.....))))))))))))......((((...(((((((...(((....)))..)))))))..))))......... ( -43.80) >DroYak_CAF1 63048 117 + 1 CCCGAUGAUCAUUGAUCAAUGAUCGUGCCUCCUGCCGUCUGG---CUACCUGCGGCACUCAGGGCCACUUAGUGAUUUUGGCCGUUGUUUGUUGACCAGGCCAAACAGCUGCCACUACUU ....(((((((((....)))))))))((((..((((((..((---...)).))))))....))))....((((...(((((((...(((....)))..)))))))..))))......... ( -38.00) >consensus CCUGAUGAUCAUUGAUCAAUGAUCGUGCCUCCUGCCGUCUGGCAGCUACCUGCGGCACUCGGGGCCACUUAGUGAUUUUGGCCGUUGUUUGUUGACCAGGCCAAACAGCUGCCACUACUU ....(((((((((....)))))))))(((((.((((((..((......)).))))))...))))).....((((....((((.((((((((..........)))))))).)))).)))). (-38.85 = -39.35 + 0.50)


| Location | 15,636,028 – 15,636,148 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -41.92 |
| Consensus MFE | -37.55 |
| Energy contribution | -38.05 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15636028 120 - 23771897 AAGUAGUGGCAGCUGUUUGGCCAGGUCAACAAACAACGGCCAAAAUCACUAAGUGGCCCCGAGUGCCGCAGGUAGCUGCCAGACGGCAGGAGGCACGAUCAUUGAUCAAUGAUCAUCAGG ..((..((((((((((((((((..((......))...)))))))........(((((.......)))))...))))))))).)).((.....))..(((((((....)))))))...... ( -43.10) >DroSec_CAF1 58136 120 - 1 AAGUAGUGGCAGCUGUUUGGCAUGGUCAACAAACAACGGCCAAAAUCACUAAGUGGACCCGAGUGCCGCAGGUAGCUGCCAGACGGCAGGAGGCACGAUCAUUGAUCAAUGAUCAUCAGG .....(((.(.((((((((((((((((..........)))))......(((.((((((....)).))))...))).)))))))))))....).)))(((((((....)))))))...... ( -40.50) >DroSim_CAF1 59103 120 - 1 AAGUAGUGGCAGCUGUUUGGCCUGGUCAACAAACAACGGCCAAAAUCACUAAGUGGCCCCGAGUGCCGCAGGUAGCUGCCAGACGGCAGGAGGCACGAUCAUUGUUCAAUGAUCAUCAGG ..((..((((((((((((((((..((......))...)))))))........(((((.......)))))...))))))))).)).((.....))..(((((((....)))))))...... ( -43.10) >DroYak_CAF1 63048 117 - 1 AAGUAGUGGCAGCUGUUUGGCCUGGUCAACAAACAACGGCCAAAAUCACUAAGUGGCCCUGAGUGCCGCAGGUAG---CCAGACGGCAGGAGGCACGAUCAUUGAUCAAUGAUCAUCGGG .....(((.(.((((((((((.(((((..........)))))..........(((((.......))))).....)---)))))))))....).)))(((((((....)))))))...... ( -41.00) >consensus AAGUAGUGGCAGCUGUUUGGCCUGGUCAACAAACAACGGCCAAAAUCACUAAGUGGCCCCGAGUGCCGCAGGUAGCUGCCAGACGGCAGGAGGCACGAUCAUUGAUCAAUGAUCAUCAGG .....(((.(.((((((((((..((((..........))))...........(((((.......)))))........))))))))))....).)))(((((((....)))))))...... (-37.55 = -38.05 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:19 2006