
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,634,943 – 15,635,214 |
| Length | 271 |
| Max. P | 0.918827 |

| Location | 15,634,943 – 15,635,063 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -36.95 |
| Energy contribution | -36.57 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15634943 120 - 23771897 AUGCCGAACCUGAACAUGAACCUCAAUGGAAAUUAUAGCAAGUCUUGUCCAUUUAAGAGGAUUUAGGUUCGGUAGGUUCGCAUUUGUAUGCUAACAAGGGCGUGCGCUGAAUGGUCUUUA .(((((((((((((......((((((((((....((.....))....))))))...)))).))))))))))))).((((((....(((((((......))))))))).))))........ ( -36.80) >DroSec_CAF1 57167 120 - 1 AUGCCGAACCUGAACAUGAACCUCAAUGGAAAUUAUAGCAAGUCUUGUCCAUUUAAGAGGAUUUAGGUUCGGUAGGUUCGCAUUUGUAUGCUAACAAGGGCGUGCGCUGAAUGGUCUUUA .(((((((((((((......((((((((((....((.....))....))))))...)))).))))))))))))).((((((....(((((((......))))))))).))))........ ( -36.80) >DroSim_CAF1 58125 120 - 1 AUGCCGAACCUGAACAUGAACCUCAAUGGAAAUUAUAGCAAGUCUUGUCCAUUUAAGAGGAUUUAGGUUCGGUAGGUUCGCAUUUGUAUGCUAACAAGGGCGUGCGCUGAAUGGUCUUUA .(((((((((((((......((((((((((....((.....))....))))))...)))).))))))))))))).((((((....(((((((......))))))))).))))........ ( -36.80) >DroYak_CAF1 61994 120 - 1 AUGCCGAGCCUGAACAUGAACCUCAAUGGAAAUUAUAGCAAGUCUUGUCCAUUUGAGGGGAUUUAGGUUCGGUAGGUUCGCAUUUGUAUGCUAACAAGGGCGUGCGCUGAAUGGUCUUUA .((((((((((((((.....((((((((((....((.....))....)))).)))))).).))))))))))))).((((((....(((((((......))))))))).))))........ ( -40.80) >consensus AUGCCGAACCUGAACAUGAACCUCAAUGGAAAUUAUAGCAAGUCUUGUCCAUUUAAGAGGAUUUAGGUUCGGUAGGUUCGCAUUUGUAUGCUAACAAGGGCGUGCGCUGAAUGGUCUUUA .(((((((((((((......((((((((((....((.....))....))))))...)))).))))))))))))).((((((....(((((((......))))))))).))))........ (-36.95 = -36.57 + -0.38)


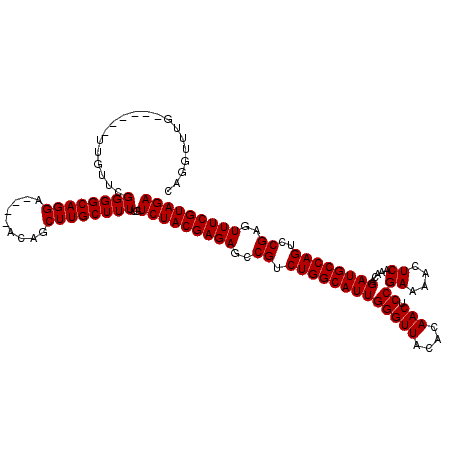
| Location | 15,634,983 – 15,635,094 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -28.94 |
| Energy contribution | -28.57 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15634983 111 - 23771897 AAGCUGUUCGUUCCUGCCCCGAAC---------AAACCUGAUGCCGAACCUGAACAUGAACCUCAAUGGAAAUUAUAGCAAGUCUUGUCCAUUUAAGAGGAUUUAGGUUCGGUAGGUUCG ....((((((.........)))))---------)(((((...((((((((((((......((((((((((....((.....))....))))))...)))).))))))))))))))))).. ( -33.70) >DroSec_CAF1 57207 110 - 1 AAGCUGU----UCCUGCCCCGAACAA------CAAACCUGAUGCCGAACCUGAACAUGAACCUCAAUGGAAAUUAUAGCAAGUCUUGUCCAUUUAAGAGGAUUUAGGUUCGGUAGGUUCG ..(.(((----((.......))))).------).(((((...((((((((((((......((((((((((....((.....))....))))))...)))).))))))))))))))))).. ( -32.10) >DroSim_CAF1 58165 110 - 1 AAGCUGU----UCCUGCCCCGAACAA------CAAACCUGAUGCCGAACCUGAACAUGAACCUCAAUGGAAAUUAUAGCAAGUCUUGUCCAUUUAAGAGGAUUUAGGUUCGGUAGGUUCG ..(.(((----((.......))))).------).(((((...((((((((((((......((((((((((....((.....))....))))))...)))).))))))))))))))))).. ( -32.10) >DroYak_CAF1 62034 116 - 1 AAGCUGU----UCCUGCCCCGAACAAACCUGAGAUGCCUGAUGCCGAGCCUGAACAUGAACCUCAAUGGAAAUUAUAGCAAGUCUUGUCCAUUUGAGGGGAUUUAGGUUCGGUAGGUUCG ....(((----((.......))))).......((.((((...(((((((((((((.....((((((((((....((.....))....)))).)))))).).)))))))))))))))))). ( -36.30) >consensus AAGCUGU____UCCUGCCCCGAACAA______CAAACCUGAUGCCGAACCUGAACAUGAACCUCAAUGGAAAUUAUAGCAAGUCUUGUCCAUUUAAGAGGAUUUAGGUUCGGUAGGUUCG ...................(((((.................(((((((((((((......((((((((((....((.....))....))))))...)))).))))))))))))).))))) (-28.94 = -28.57 + -0.37)



| Location | 15,635,063 – 15,635,174 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15635063 111 + 23771897 CAGGUUU---------GUUCGGGGCAGGAACGAACAGCUUGCUUUUGUCUACGAGAGCCGUCUGGCAUUGGGUUACACAACUCCGAAAACUCAAACAGAUGCCAGUCCGAGUUUCGUAGA ((((((.---------(((((.........)))))))))))......(((((((((..((.(((((((((((((....))).))((....)).....))))))))..))..))))))))) ( -37.40) >DroSec_CAF1 57287 110 + 1 CAGGUUUG------UUGUUCGGGGCAGGA----ACAGCUUGCUUUUGUCUACGAGAGCCGUCUGGCAUUGGGUUACACAACUCCGAAAACUCAAACAGAUGCCAGUCCGAGUUUCGUAGA .((((..(------((((((.......))----)))))..))))...(((((((((..((.(((((((((((((....))).))((....)).....))))))))..))..))))))))) ( -38.10) >DroSim_CAF1 58245 110 + 1 CAGGUUUG------UUGUUCGGGGCAGGA----ACAGCUUGCUUUUGUCUACGAGAGCCGUCUGGCAUUGGGUUACACAACUCCGAAAACUCAAACAGAUGCCAGUCCGAGUUUCGUAGA .((((..(------((((((.......))----)))))..))))...(((((((((..((.(((((((((((((....))).))((....)).....))))))))..))..))))))))) ( -38.10) >DroYak_CAF1 62114 116 + 1 CAGGCAUCUCAGGUUUGUUCGGGGCAGGA----ACAGCUUGCUUUUGUCUACGAGAGCCGUCUGGCAUUGGGUUACACAACUCCGAAAACUCAAACAGAUGCCAGUCCGAGUUUCGUAGA (((((.......)))))...((((((((.----....))))))))..(((((((((..((.(((((((((((((....))).))((....)).....))))))))..))..))))))))) ( -35.60) >consensus CAGGUUUG______UUGUUCGGGGCAGGA____ACAGCUUGCUUUUGUCUACGAGAGCCGUCUGGCAUUGGGUUACACAACUCCGAAAACUCAAACAGAUGCCAGUCCGAGUUUCGUAGA ....................((((((((.........))))))))..(((((((((..((.(((((((((((((....))).))((....)).....))))))))..))..))))))))) (-33.00 = -33.00 + 0.00)


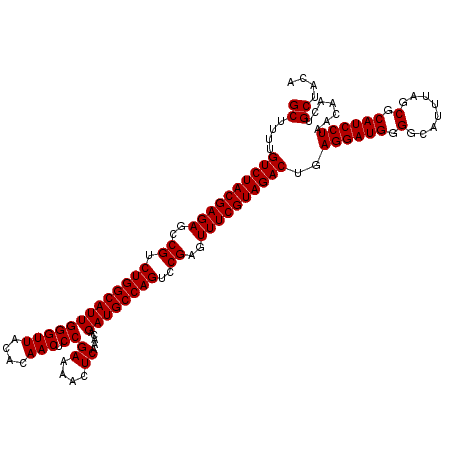
| Location | 15,635,094 – 15,635,214 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -40.80 |
| Energy contribution | -40.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15635094 120 + 23771897 GCUUUUGUCUACGAGAGCCGUCUGGCAUUGGGUUACACAACUCCGAAAACUCAAACAGAUGCCAGUCCGAGUUUCGUAGACUGAGGAUGGGGCAUUUAGCGCAUCCUAACAACUGCUACA ((....((((((((((..((.(((((((((((((....))).))((....)).....))))))))..))..))))))))))..((((((.(........).)))))).......)).... ( -40.90) >DroSec_CAF1 57317 120 + 1 GCUUUUGUCUACGAGAGCCGUCUGGCAUUGGGUUACACAACUCCGAAAACUCAAACAGAUGCCAGUCCGAGUUUCGUAGACUGAGGAUGGGGCAUUUAGCGCAUCCUAACAACUGCUACA ((....((((((((((..((.(((((((((((((....))).))((....)).....))))))))..))..))))))))))..((((((.(........).)))))).......)).... ( -40.90) >DroSim_CAF1 58275 120 + 1 GCUUUUGUCUACGAGAGCCGUCUGGCAUUGGGUUACACAACUCCGAAAACUCAAACAGAUGCCAGUCCGAGUUUCGUAGACUGAGGAUGGGGCAUUUAGCGCAUCCUAACAACUGCUACA ((....((((((((((..((.(((((((((((((....))).))((....)).....))))))))..))..))))))))))..((((((.(........).)))))).......)).... ( -40.90) >DroYak_CAF1 62150 120 + 1 GCUUUUGUCUACGAGAGCCGUCUGGCAUUGGGUUACACAACUCCGAAAACUCAAACAGAUGCCAGUCCGAGUUUCGUAGACCGAGGAUGGGGCAUUUAGCGCAUCCUAACAACUGCUACA ((....((((((((((..((.(((((((((((((....))).))((....)).....))))))))..))..))))))))))..((((((.(........).)))))).......)).... ( -40.50) >consensus GCUUUUGUCUACGAGAGCCGUCUGGCAUUGGGUUACACAACUCCGAAAACUCAAACAGAUGCCAGUCCGAGUUUCGUAGACUGAGGAUGGGGCAUUUAGCGCAUCCUAACAACUGCUACA ((....((((((((((..((.(((((((((((((....))).))((....)).....))))))))..))..))))))))))..((((((.(........).)))))).......)).... (-40.80 = -40.80 + 0.00)


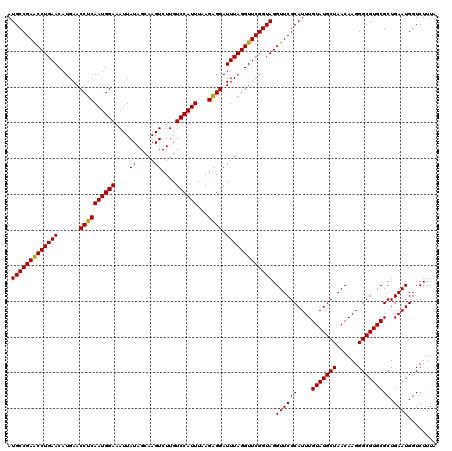
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:16 2006