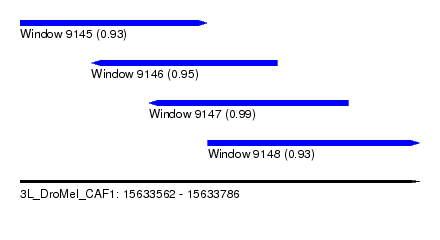
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,633,562 – 15,633,786 |
| Length | 224 |
| Max. P | 0.991182 |

| Location | 15,633,562 – 15,633,667 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.43 |
| Mean single sequence MFE | -20.27 |
| Consensus MFE | -18.88 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15633562 105 + 23771897 CGGGGAUCGCAUAAAAAUGAUUUCGUUUUGAUUGCAAUAACUCCCCCAAUUUUCUUGGCUAAUCCCCCACCA--------------CCCGCCGCCCCACCCACACCCCUU-UUUGGAGAU .((((((.(((..((((((....))))))...)))..........((((.....))))...)))))).....--------------....................((..-...)).... ( -20.20) >DroSec_CAF1 55831 104 + 1 CGGGGAUCGCAUAAAAAUGAUUUCGUUUUGAUUGCCAUAACUCCCCCAAUUUUCUUGGCUAAUCCCCCACCG--------------CCCGCUGCCCCACCCACACCCCUU-UUU-GAGAU .((((...((...((((((....))))))((((((((..................)))).))))........--------------...))..)))).............-...-..... ( -20.27) >DroSim_CAF1 56789 104 + 1 CGGGGAUCGCAUAAAAAUGAUUUCGUUUUGAUUGCAAUAACUCCCCCAAUUUUCUUGGCUAAUCCCCCACCA--------------CCCGCUGCCCCACCCACACCCCUU-UUU-GAGAU .((((((.(((..((((((....))))))...)))..........((((.....))))...)))))).....--------------........................-...-..... ( -19.10) >DroYak_CAF1 60581 119 + 1 CGGGGAUCGCAUAAAAAUGAUUUCGUUUUGAUUGCAAUAACUCCCCCAAUUUUCUUGGCUAAUCCCCCACUCCCCACCCCGCAUCACGCACUGCCCCACCCACACCCUUUGUUU-GAGAU .((((((.(((..((((((....))))))...)))..........((((.....))))...))))))..(((........((.....))............(((.....)))..-))).. ( -21.50) >consensus CGGGGAUCGCAUAAAAAUGAUUUCGUUUUGAUUGCAAUAACUCCCCCAAUUUUCUUGGCUAAUCCCCCACCA______________CCCGCUGCCCCACCCACACCCCUU_UUU_GAGAU .((((((.(((..((((((....))))))...)))..........((((.....))))...))))))..................................................... (-18.88 = -18.88 + 0.00)

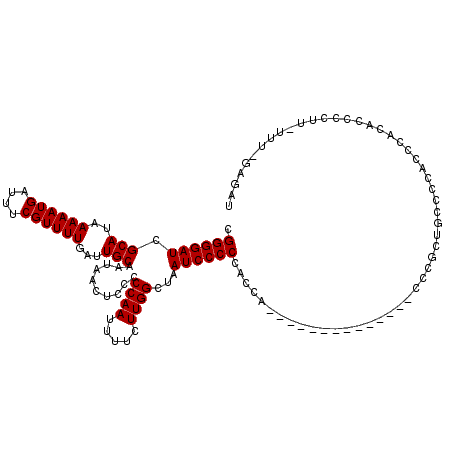

| Location | 15,633,602 – 15,633,706 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.80 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -30.14 |
| Energy contribution | -30.20 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15633602 104 - 23771897 U-UCGGGGGUAGAACAACCCUCAAGUAGUGCACUUGCCACAUCUCCAAA-AAGGGGUGUGGGUGGGGCGGCGGG--------------UGGUGGGGGAUUAGCCAAGAAAAUUGGGGGAG .-...............(((((((.....((.((..(((((((((....-..))))))).))..))))(((..(--------------(........))..))).......))))))).. ( -34.60) >DroSec_CAF1 55871 104 - 1 UUUUGGGGGUAGAACAACCCUCAAGUAGUGCACUUGCCACAUCUC-AAA-AAGGGGUGUGGGUGGGGCAGCGGG--------------CGGUGGGGGAUUAGCCAAGAAAAUUGGGGGAG ((((((.(......)..((((((.((.((((.((..(((((((((-...-..))))))).))..))))).)..)--------------)..)))))).....))))))............ ( -35.80) >DroSim_CAF1 56829 104 - 1 UUUUGGGGGUAGAACAACCCUCAAGUAGUGCACUUGCCACAUCUC-AAA-AAGGGGUGUGGGUGGGGCAGCGGG--------------UGGUGGGGGAUUAGCCAAGAAAAUUGGGGGAG .(((((((((......)))))))))...(((.((..(((((((((-...-..))))))).))..)))))..((.--------------((((.....)))).))................ ( -35.50) >DroYak_CAF1 60621 116 - 1 UU---GGGGUAGAACAACCCUCAAGUAGUGCACUUGCCACAUCUC-AAACAAAGGGUGUGGGUGGGGCAGUGCGUGAUGCGGGGUGGGGAGUGGGGGAUUAGCCAAGAAAAUUGGGGGAG ..---............((((((((((.(((.((..((((((((.-.......)))))).))..))))).)))......(..(((...((........)).)))..)....))))))).. ( -35.80) >consensus UUUUGGGGGUAGAACAACCCUCAAGUAGUGCACUUGCCACAUCUC_AAA_AAGGGGUGUGGGUGGGGCAGCGGG______________UGGUGGGGGAUUAGCCAAGAAAAUUGGGGGAG .................(((((((....(((.((..(((((((((.......))))))).))..)))))......................(((........)))......))))))).. (-30.14 = -30.20 + 0.06)

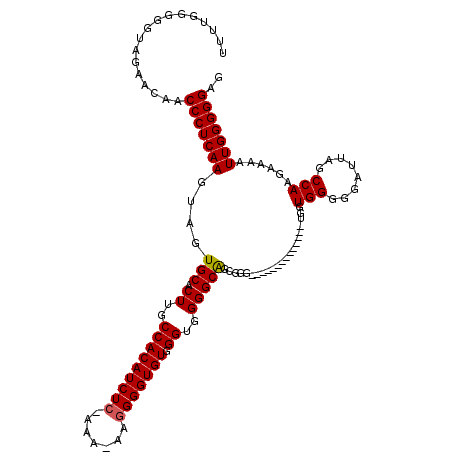
| Location | 15,633,634 – 15,633,746 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -38.96 |
| Energy contribution | -39.90 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15633634 112 - 23771897 AGCUACAAACAAUUGGCAACACUUGUGUUGUGCGGCGAUUU-UCGGGGGUAGAACAACCCUCAAGUAGUGCACUUGCCACAUCUCCAAA-AAGGGGUGUGGGUGGGGCGGCGGG------ .((.((((((((.((....)).)))).)))))).((.....-..((((((......))))))......(((.((..(((((((((....-..))))))).))..)))))))...------ ( -41.30) >DroSec_CAF1 55903 112 - 1 AGCUACAAACAAUUGGCAACACUUGUGUUGUGCGGCGAUUUUUUGGGGGUAGAACAACCCUCAAGUAGUGCACUUGCCACAUCUC-AAA-AAGGGGUGUGGGUGGGGCAGCGGG------ .(((((((((((.((....)).)))).))))((.(((....(((((((((......)))))))))...))).((..(((((((((-...-..))))))).))..)))))))...------ ( -44.50) >DroSim_CAF1 56861 112 - 1 AGCUACAAACAAUUGGCAACACUUGUGUUGUGCGGCGAUUUUUUGGGGGUAGAACAACCCUCAAGUAGUGCACUUGCCACAUCUC-AAA-AAGGGGUGUGGGUGGGGCAGCGGG------ .(((((((((((.((....)).)))).))))((.(((....(((((((((......)))))))))...))).((..(((((((((-...-..))))))).))..)))))))...------ ( -44.50) >DroYak_CAF1 60661 116 - 1 AGCUACAAACAAUUGGCAACACUUGUGUUGUGCGGCGACUUU---GGGGUAGAACAACCCUCAAGUAGUGCACUUGCCACAUCUC-AAACAAAGGGUGUGGGUGGGGCAGUGCGUGAUGC .((.((((((((.((....)).)))).)))))).((.((((.---(((((......))))).)))).))(((((.(((.((((.(-(.((.....)).)))))).))))))))....... ( -41.30) >consensus AGCUACAAACAAUUGGCAACACUUGUGUUGUGCGGCGAUUUUUUGGGGGUAGAACAACCCUCAAGUAGUGCACUUGCCACAUCUC_AAA_AAGGGGUGUGGGUGGGGCAGCGGG______ .(((((((((((.((....)).)))).))))((.((.(((.(((((((((......))))))))).))))).((..(((((((((.......))))))).))..)))))))......... (-38.96 = -39.90 + 0.94)

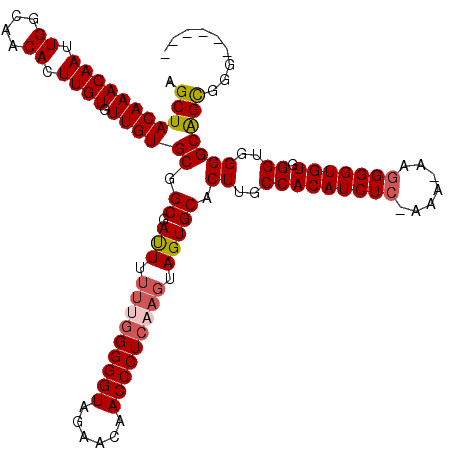
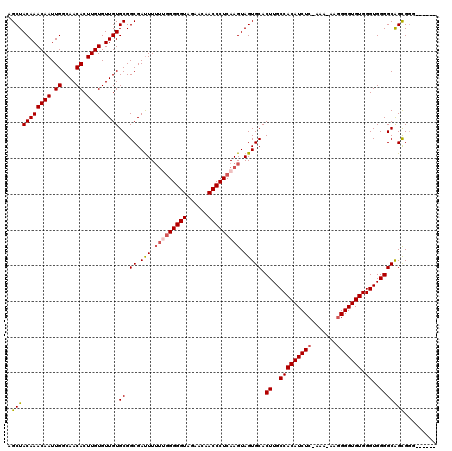
| Location | 15,633,667 – 15,633,786 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.79 |
| Mean single sequence MFE | -37.57 |
| Consensus MFE | -35.33 |
| Energy contribution | -35.33 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15633667 119 + 23771897 GUGGCAAGUGCACUACUUGAGGGUUGUUCUACCCCCGA-AAAUCGCCGCACAACACAAGUGUUGCCAAUUGUUUGUAGCUAUUUUGUCCGGCGACAGCCUCCCAGAAAUUGGCAUCACAA ((((((((((...)))))(.((((......)))).)..-.....))))).........(((.((((((((.((((..((......))..(((....)))...)))))))))))).))).. ( -36.00) >DroSec_CAF1 55935 119 + 1 GUGGCAAGUGCACUACUUGAGGGUUGUUCUACCCCCAAAAAAUCGCCGCACAACACAAGUGUUGCCAAUUGUUUGUAGCUAUUUUGUC-GGCGACAGCCUCCCAGAAAUUGGCAUCACAA (((((.((....))..(((.((((......)))).)))......))))).........(((.((((((((.((((..((......)).-(((....)))...)))))))))))).))).. ( -36.10) >DroSim_CAF1 56893 119 + 1 GUGGCAAGUGCACUACUUGAGGGUUGUUCUACCCCCAAAAAAUCGCCGCACAACACAAGUGUUGCCAAUUGUUUGUAGCUAUUUUGUC-GGCGACAGCCUCCCAGAAAUUGGCAUCACAA (((((.((....))..(((.((((......)))).)))......))))).........(((.((((((((.((((..((......)).-(((....)))...)))))))))))).))).. ( -36.10) >DroYak_CAF1 60700 116 + 1 GUGGCAAGUGCACUACUUGAGGGUUGUUCUACCCC---AAAGUCGCCGCACAACACAAGUGUUGCCAAUUGUUUGUAGCUAUUUGGUC-GGCGACAGCCUCCCAGAAAUUGGCAUCACAA (((....((((.(.((((..((((......)))).---.)))).)..))))..)))..(((.((((((((((.....))..(((((..-(((....)))..))))))))))))).))).. ( -42.10) >consensus GUGGCAAGUGCACUACUUGAGGGUUGUUCUACCCCCAAAAAAUCGCCGCACAACACAAGUGUUGCCAAUUGUUUGUAGCUAUUUUGUC_GGCGACAGCCUCCCAGAAAUUGGCAUCACAA ((((((((((...)))))..((((......))))..........))))).........(((.((((((((.((((..((......))..(((....)))...)))))))))))).))).. (-35.33 = -35.33 + -0.00)

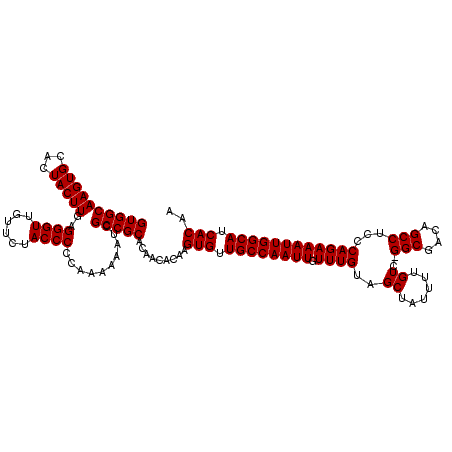
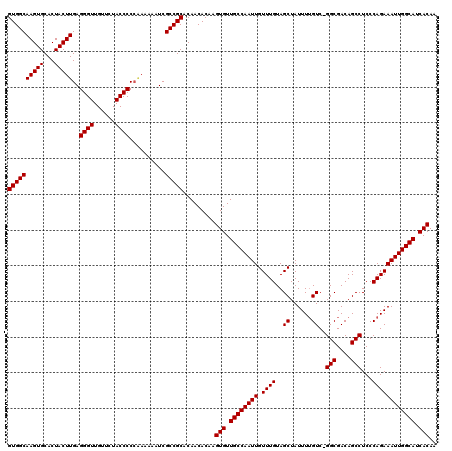
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:10 2006