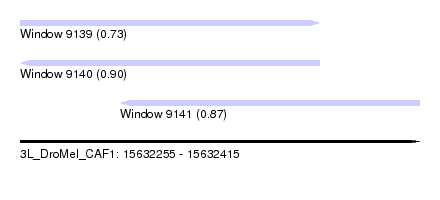
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 15,632,255 – 15,632,415 |
| Length | 160 |
| Max. P | 0.896872 |

| Location | 15,632,255 – 15,632,375 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -23.48 |
| Energy contribution | -22.97 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15632255 120 + 23771897 AUAAGCUGACCCAGUUUUGGAUUCAUAUUUUGAUACAAAUUGGUUGCAACAUAUCCUGAAAGUCAAACUUUCUGGACCGCUUCAGAUAACCACAAUUAAAUGGCAACAUAAUAUUGGGCA ..((((((..(((((((.(...(((.....)))..))))))))...)).....(((.(((((.....))))).)))..)))).......((.((((...(((....)))...)))))).. ( -26.50) >DroSec_CAF1 54505 120 + 1 AUAAGCUGACCCAGUUUUGGAUUCAUAUUUUGAUACAAAUUGGUUGCAACAUUUCCUGAAAGUCAAACUUUCUGGCCUGUGUCAGAUUACCACAAUUAAAUGGCAACCUAAUAUUGGGCA .......(.((((((((((...(((.....)))..))))..((((((..(((((((.(((((.....))))).))..((((.........))))...)))))))))))....))))))). ( -28.10) >DroSim_CAF1 55461 120 + 1 AUAAGCUGACCCAGUUUUGGAUUCAUAUUUUGAUACAAAUUGGUUGCAACAUUUCCUGAAAGUCAAACUUUUUGGCCUUCGUCAGAUUACUACAAUUAAAUGGCAACCUAAUAUUGGGCA .......(.((((((((((...(((.....)))..))))..((((((..(((((...(((.(((((.....))))).)))((.((....))))....)))))))))))....))))))). ( -25.60) >DroYak_CAF1 59145 120 + 1 AUAAGCUGACCCAGUUUUGGAUUCAUAUCUUGAUACAAAUUGGUUGCAACAUUUCAUGAAAGUCAAACUUUCUGGGCUGCUUCAGAUUACCACAAUUAAAUGGCAACCUAAUAUUGGGCA .......(.((((((((((...(((.....)))..))))..((((((..(((((.(((((((.....)))))(((.(((...)))....)))..)).)))))))))))....))))))). ( -27.50) >consensus AUAAGCUGACCCAGUUUUGGAUUCAUAUUUUGAUACAAAUUGGUUGCAACAUUUCCUGAAAGUCAAACUUUCUGGCCUGCGUCAGAUUACCACAAUUAAAUGGCAACCUAAUAUUGGGCA .......(.((((((((((...(((.....)))..))))..((((((..........(((((.....)))))((((....))))..................))))))....))))))). (-23.48 = -22.97 + -0.50)


| Location | 15,632,255 – 15,632,375 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -26.10 |
| Energy contribution | -26.85 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15632255 120 - 23771897 UGCCCAAUAUUAUGUUGCCAUUUAAUUGUGGUUAUCUGAAGCGGUCCAGAAAGUUUGACUUUCAGGAUAUGUUGCAACCAAUUUGUAUCAAAAUAUGAAUCCAAAACUGGGUCAGCUUAU .(((((((...(((....)))...)))).)))......(((((..((((((((.....))))).((((....(((((.....)))))(((.....))))))).....)))..).)))).. ( -26.30) >DroSec_CAF1 54505 120 - 1 UGCCCAAUAUUAGGUUGCCAUUUAAUUGUGGUAAUCUGACACAGGCCAGAAAGUUUGACUUUCAGGAAAUGUUGCAACCAAUUUGUAUCAAAAUAUGAAUCCAAAACUGGGUCAGCUUAU .((......((((((((((((......))))))))))))....((((....(((((....((((.....((.(((((.....))))).)).....))))....))))).)))).)).... ( -31.60) >DroSim_CAF1 55461 120 - 1 UGCCCAAUAUUAGGUUGCCAUUUAAUUGUAGUAAUCUGACGAAGGCCAAAAAGUUUGACUUUCAGGAAAUGUUGCAACCAAUUUGUAUCAAAAUAUGAAUCCAAAACUGGGUCAGCUUAU .((......(((((((((.((......)).)))))))))....((((....(((((....((((.....((.(((((.....))))).)).....))))....))))).)))).)).... ( -25.10) >DroYak_CAF1 59145 120 - 1 UGCCCAAUAUUAGGUUGCCAUUUAAUUGUGGUAAUCUGAAGCAGCCCAGAAAGUUUGACUUUCAUGAAAUGUUGCAACCAAUUUGUAUCAAGAUAUGAAUCCAAAACUGGGUCAGCUUAU ..........(((((((((((......)))))))))))((((.((((((....((((...((((((...((.(((((.....))))).))...))))))..)))).))))))..)))).. ( -35.50) >consensus UGCCCAAUAUUAGGUUGCCAUUUAAUUGUGGUAAUCUGAAGCAGGCCAGAAAGUUUGACUUUCAGGAAAUGUUGCAACCAAUUUGUAUCAAAAUAUGAAUCCAAAACUGGGUCAGCUUAU .((((....((((((((((((......))))))))))))............(((((....((((.....((.(((((.....))))).)).....))))....)))))))))........ (-26.10 = -26.85 + 0.75)



| Location | 15,632,295 – 15,632,415 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -27.59 |
| Energy contribution | -28.15 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15632295 120 - 23771897 CGGAACAAAGGGGAGUUUAAACUUUUAAGUGGAAGAGAAUUGCCCAAUAUUAUGUUGCCAUUUAAUUGUGGUUAUCUGAAGCGGUCCAGAAAGUUUGACUUUCAGGAUAUGUUGCAACCA .(.((((..(((.(((((...(((((....))))).))))).)))....(((.((.(((((......))))).)).)))....((((.(((((.....))))).)))).)))).)..... ( -28.60) >DroSec_CAF1 54545 120 - 1 CGGAACAAAGGGGAGUUUAAACUUUUAAGUGGAAGAGAAUUGCCCAAUAUUAGGUUGCCAUUUAAUUGUGGUAAUCUGACACAGGCCAGAAAGUUUGACUUUCAGGAAAUGUUGCAACCA .((......(((.(((((...(((((....))))).))))).)))....((((((((((((......)))))))))))).(((..((.(((((.....))))).))...))).....)). ( -34.30) >DroSim_CAF1 55501 120 - 1 CGGAACAAAGGGGAGUUUAAACUUUUAAGUGGAAGAGAAUUGCCCAAUAUUAGGUUGCCAUUUAAUUGUAGUAAUCUGACGAAGGCCAAAAAGUUUGACUUUCAGGAAAUGUUGCAACCA .(.((((..(((.(((((...(((((....))))).))))).)))....(((((((((.((......)).))))))))).(((((.(((.....))).)))))......)))).)..... ( -26.80) >DroYak_CAF1 59185 120 - 1 CGGAACAAAAGGGAGCUUAAACUUUUAAGUGGAACAGAAUUGCCCAAUAUUAGGUUGCCAUUUAAUUGUGGUAAUCUGAAGCAGCCCAGAAAGUUUGACUUUCAUGAAAUGUUGCAACCA .((.......(((.((((((....))))))............)))....((((((((((((......)))))))))))).(((((.(((((((.....))))).))....)))))..)). ( -33.22) >consensus CGGAACAAAGGGGAGUUUAAACUUUUAAGUGGAAGAGAAUUGCCCAAUAUUAGGUUGCCAUUUAAUUGUGGUAAUCUGAAGCAGGCCAGAAAGUUUGACUUUCAGGAAAUGUUGCAACCA .(.((((...(..((.((((((((((...(((...........)))...((((((((((((......)))))))))))).........))))))))))))..)......)))).)..... (-27.59 = -28.15 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:04 2006