
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
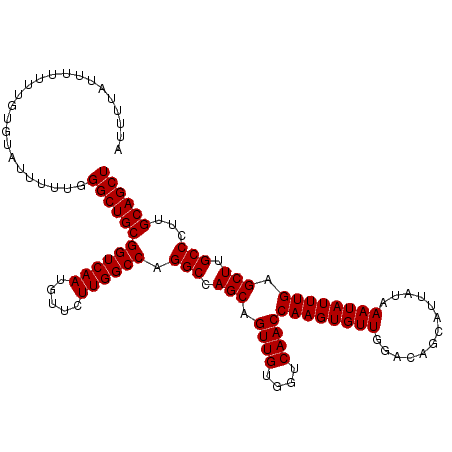
| Location | 15,631,946 – 15,632,055 |
| Length | 109 |
| Max. P | 0.992796 |

| Location | 15,631,946 – 15,632,055 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 95.32 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -32.85 |
| Energy contribution | -32.85 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15631946 109 + 23771897 AUUU-----UCUUGUGUAUUUUUUGGCUGCGGUCAAUGUUCUUGGCCAGGCCAGCAGUUGUGGUCAACCAAGUGUUGGACAGCAUUAUAAAUAUUUGAGCUUGCCCUUGCAGCU ....-----...............((((((((((((.....)))))).(((.(((.((((....))))((((((((.............)))))))).))).)))...)))))) ( -32.72) >DroSec_CAF1 54191 114 + 1 AUUUUAUUUUUUUGUGUAUUUUUGGGCUGCGGUCAAUGUUCUUGGCCAGGCCAGCAGUUGUGGUCAACCAAGUGUUGGACAGCAUUAUAAAUAUUUGAGCUUGCCCUUGCAGCU ........................((((((((((((.....)))))).(((.(((.((((....))))((((((((.............)))))))).))).)))...)))))) ( -32.92) >DroSim_CAF1 55148 113 + 1 AUUUUAUUU-UUUGUGUAUUUUUGGGCUGCGGUCAAUGUUCUUGGCCAGGCCAGCAGUUGUGGUCAACCAAGUGUUGGACAGCAUUAUAAAUAUUUGAGCUUGCCCUUGCAGCU .........-..............((((((((((((.....)))))).(((.(((.((((....))))((((((((.............)))))))).))).)))...)))))) ( -32.92) >consensus AUUUUAUUUUUUUGUGUAUUUUUGGGCUGCGGUCAAUGUUCUUGGCCAGGCCAGCAGUUGUGGUCAACCAAGUGUUGGACAGCAUUAUAAAUAUUUGAGCUUGCCCUUGCAGCU ........................((((((((((((.....)))))).(((.(((.((((....))))((((((((.............)))))))).))).)))...)))))) (-32.85 = -32.85 + 0.00)

| Location | 15,631,946 – 15,632,055 |
|---|---|
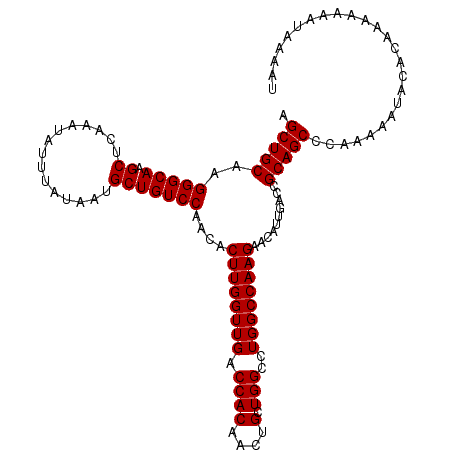
| Length | 109 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 95.32 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -27.66 |
| Energy contribution | -27.66 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.82 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 15631946 109 - 23771897 AGCUGCAAGGGCAAGCUCAAAUAUUUAUAAUGCUGUCCAACACUUGGUUGACCACAACUGCUGGCCUGGCCAAGAACAUUGACCGCAGCCAAAAAAUACACAAGA-----AAAU .(((((..(((((.((...............)))))))....((((((((.((((....).)))..))))))))..........)))))................-----.... ( -27.66) >DroSec_CAF1 54191 114 - 1 AGCUGCAAGGGCAAGCUCAAAUAUUUAUAAUGCUGUCCAACACUUGGUUGACCACAACUGCUGGCCUGGCCAAGAACAUUGACCGCAGCCCAAAAAUACACAAAAAAAUAAAAU .(((((..(((((.((...............)))))))....((((((((.((((....).)))..))))))))..........)))))......................... ( -27.66) >DroSim_CAF1 55148 113 - 1 AGCUGCAAGGGCAAGCUCAAAUAUUUAUAAUGCUGUCCAACACUUGGUUGACCACAACUGCUGGCCUGGCCAAGAACAUUGACCGCAGCCCAAAAAUACACAAA-AAAUAAAAU .(((((..(((((.((...............)))))))....((((((((.((((....).)))..))))))))..........)))))...............-......... ( -27.66) >consensus AGCUGCAAGGGCAAGCUCAAAUAUUUAUAAUGCUGUCCAACACUUGGUUGACCACAACUGCUGGCCUGGCCAAGAACAUUGACCGCAGCCCAAAAAUACACAAAAAAAUAAAAU .(((((..(((((.((...............)))))))....((((((((.((((....).)))..))))))))..........)))))......................... (-27.66 = -27.66 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:59:01 2006